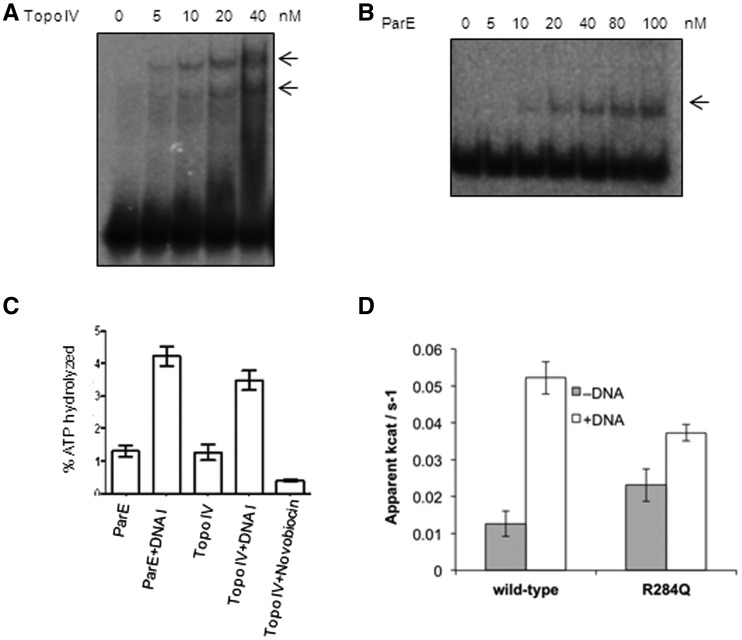
Figure 6.
DNA binding and ATPase activity of E. coli topoIV and its ParE subunit (A) EMSA with E. coli topo IV holoenzyme (5–40 nM) using 72-bp DNA. The arrows indicate two DNA-bound complexes; (B) EMSA with increasing concentrations of ParE (5–100 nM) subunit using 72-bp DNA. The reaction conditions were similar to those used for Figure 1; (C) ATP hydrolysis by the topo IV holoenzyme and ParE (100 nM of each) subunit in the absence and presence of 72-bp (100 nM) DNA; (D) ATPase activity of wild-type and R284Q mutant ParE43 fragment (2 μM dimer), in the presence and absence of linear pBR322 at a ratio of 25-bp DNA per protein dimer. The activity is expressed as the apparent kcat of the protein dimer in the presence of 2 mM ATP. The error bars represent the standard deviation of three measurements.