Table 1.
The apparent equilibrium and kinetic binding parameters between H/ACA RNPs and substrate RNAs at 27°C
| Substrate | H/ACA RNP |
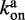 (103 M−1 s−1) (103 M−1 s−1) |
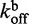 (10−3 s−1) (10−3 s−1) |
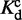 (µM) (µM) |
|---|---|---|---|---|
| Sub-Ud | WT | 27 ± 1 | n.d. | n.d. |
| Sub-U | D85A | 17 ± 1 | 0.69 ± 0.09 | 0.031 ± 0.002 |
| Sub-U | DEL7 | 18 ± 3 | 12 ± 3 | 0.23 ± 0.08 |
| Sub-U | R154Q | 11 ± 3 | 11 ± 2 | 0.47 ± 0.05 |
| Sub-U | RNA only | n.d. | 27 ± 5 | 1.5 ± 0.1 |
| Sub-Ψ | WT | 23 ± 5 | 12 ± 1 | 1.1 ± 0.2 |
| Sub-Ψ | D85A | 22 ± 4 | 11 ± 1 | 0.9 ± 0.1 |
| Sub-Ψ | DEL7 | 15 ± 3 | 14 ± 1 | 0.9 ± 0.1 |
| Sub-Ψ | R154Q | 18 ± 8 | 16 ± 2 | 0.8 ± 0.1 |
| Sub-Ψ | RNA only | n.d. | 17 ± 5 | 1.72 ± 0.15 |
| Sub-Ud | ΔGar1 | 12 ± 1 | n.d. | n.d. |
| Sub-U | ΔGar1/D85A | 26 ± 1 | 0.54 ± 0.04 | 0.036 ± 0.002 |
| Sub-U | ΔGar1/DEL7 | 8.4 ± 0.4 | 7.8 ± 0.7 | 0.15 ± 0.01 |
| Sub-U | ΔGar1/R154Q | 8 ± 2 | 7 ± 1 | 0.42 ± 0.06 |
| Sub-Ψ | ΔGar1 | 27 ± 5 | 5 ± 1 | 0.32 ± 0.04 |
| Sub-Ψ | ΔGar1/D85A | 35 ± 4 | 5 ± 1 | 0.20 ± 0.03 |
| Sub-Ψ | ΔGar1/DEL7 | 22 ± 3 | 12 ± 1 | 0.7 ± 0.1 |
| Sub-Ψ | ΔGar1/R154Q | 24 ± 11 | 16 ± 4 | 0.83 ± 0.09 |
| Sub-C | WT | n.d. | 42 ± 9 | 3.5 ± 0.1 |
aThe errors of kon are from fitting.
bkoff values are the mean ± SD from 2–3 independent measurements.
cKd values are the mean ± SD from 2–3 independent measurements.
dThese substrate–enzyme combinations are reactive. Due to the fact that the concentrations of substrate and product RNAs vary with time, the apparent Kd and koff would be time dependent and, therefore, cannot be determined by conventional analysis, n. d., not determined.
