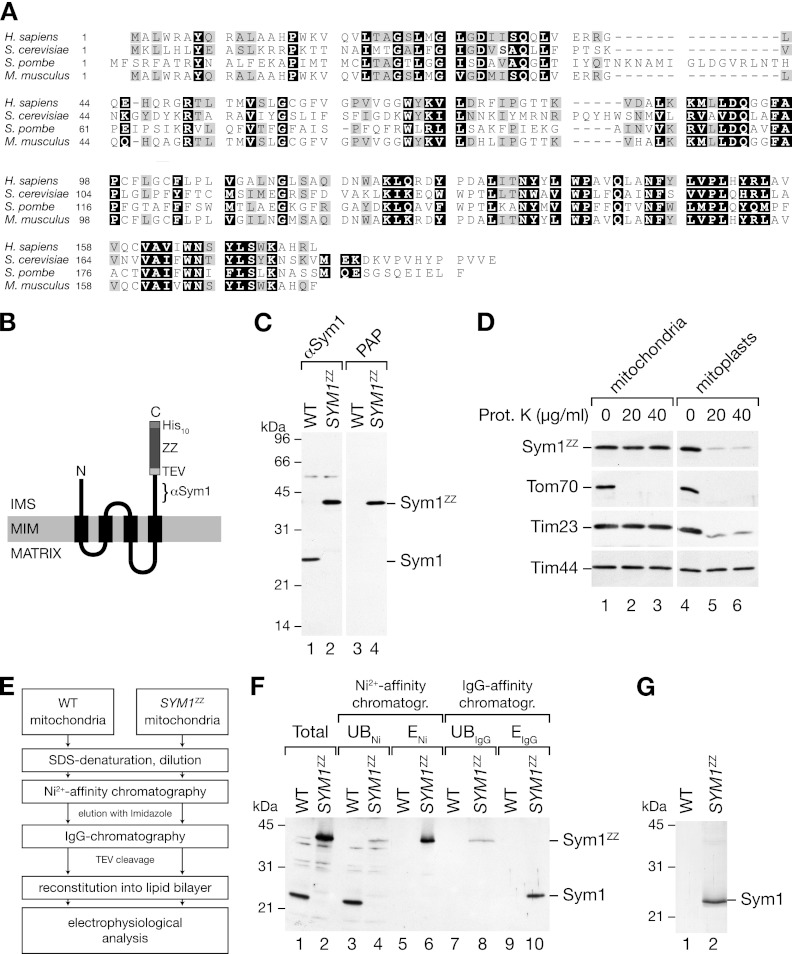
Fig 1.
Isolation of Sym1 from mitochondrial membranes. (A) ClustalW2 sequence alignment of Sym1 homologues in fungi and mammals. Black boxes indicate 100% similarity, and gray boxes indicate 75% similarity. (B) Schematic representation of the ZZ tag fused to Sym1 (ZZ, two repeats of the Z domain of protein A; TEV, cleavage site of the tobacco etch virus protease; His10, decahistidine tag). The epitope recognized by the Sym1 antibody is indicated (αSym1). IMS, intermembrane space; MIM, mitochondrial inner membrane. (C) Isolated mitochondria from the wild type (WT) and the SYM1ZZ strain were subjected to SDS-PAGE and Western blotting using antibodies against Sym1 and PAP (HRP–anti-HRP). (D) Mitochondria were left untreated or subjected to swelling in hypo-osmotic buffer, treated with the indicated amounts of protease, and subjected to SDS-PAGE and Western blotting. Sym1ZZ was detected using PAP. Prot. K, proteinase K. (E) Flow diagram of the isolation strategy for Sym1ZZ. (F) Total (5%), unbound (UB) (5%), and eluate (E) (ENi, 5%; EIgG, 100%) fractions of Ni-NTA affinity and IgG affinity chromatography were separated by SDS-PAGE and subjected to Western blot analyses using the Sym1 antibody. (G) Coomassie brilliant blue-stained SDS-PAGE gel of the IgG chromatography eluate.