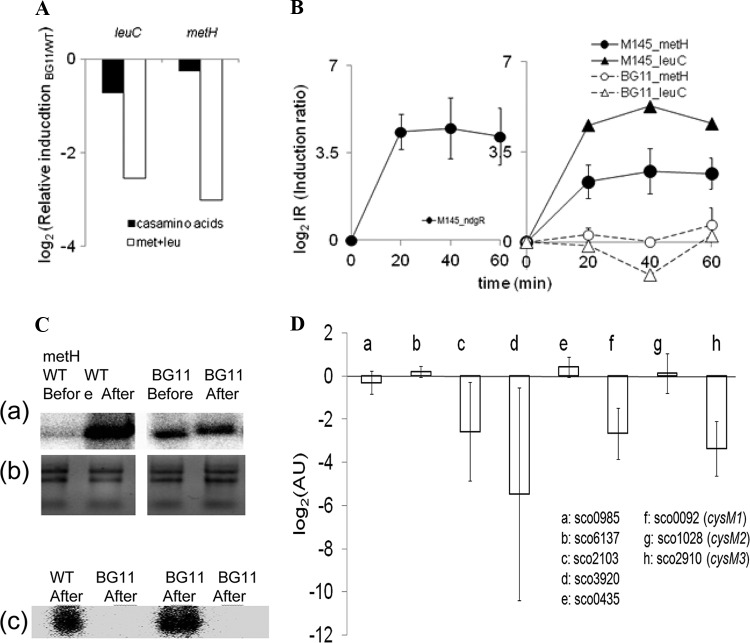
Fig 4.
NdgR-dependent gene expression. (A) Expression of leuC and metH was reduced in BG11 compared to M145 (y axis. Real-time qRT-PCR was performed using cells grown in minimal medium supplemented with Casamino Acids or Met-Leu (at 37 μg/ml). (B) Relative fold changes were compared, before and after medium exchange (log2 relative induction ratio of fold change after exchange/fold change before exchange. Instant induction of ndgR was noted (left). The mRNA expression of leuC-metH increased upon nutrient downshift in the wild type (filled symbols) but not in BG11 (open symbols) (right). (C) Low-resolution S1 mapping for metH and metF. Expression of metH was examined, comparing before and after nutrient downshift (panel a). The integrity of total RNA used in the assay was examined (panel b). A dramatic difference in metH and metF expression upon nutrient downshift was observed (panel c). RNA samples from 40 min after nutrient downshift were used. (D) The relative gene expression of BG11 over M145 by medium exchange was compared (AU, arbitrary units; log2 scale). The relative induction level (induction at 40 min/induction at time zero) was calculated for each strain and used in comparing the expression in BG11 with that in M145 (AU, in log2 scale). Error bars represent standard deviations of three independent experiments.