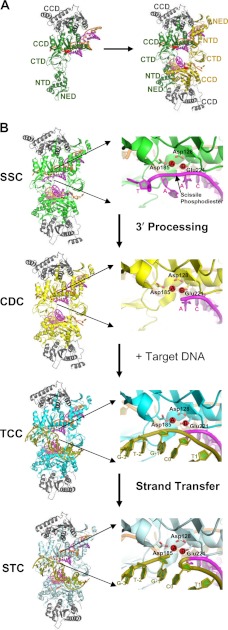
FIGURE 2.
PFV intasome structures. A, left, crystallographic asymmetric unit with the IN dimer bound to a single viral DNA end (Protein Data Bank (PDB) code 3OY9) (18, 68) and different IN domains labeled. The inner monomer is colored green, the outer monomer gray, and the DNA strands magenta (the transferred or to-be-integrated strand) and orange (the non-transferred strand). Right, two dimers come together to form the functional CDC (with the second inner monomer shown in gold). CTD, C-terminal domain; NED, N-terminal extension domain. B, left, structures of the PFV intasome complexes along the integration pathway (SSC, PDB code 4E7I; CDC, same as in A; TCC, PDB code 4E7K; and STC, PDB code 4E7L) (13). The coloring of the inner monomers matches the analogous complexes in Fig. 1. Right, enlargements of the active site of the upper inner monomer to the left, with Mn2+ ions A and B and the oxygen atoms of active site residues Asp-128, Asp-185, and Glu-221 colored red and the target DNA strand colored olive green (other coloring as described for A). The scissile viral DNA phosphodiester adjacent to the invariant CA dinucleotide and terminal residues of the transferred LTR strand in the active site regions of the SSC and CDC are labeled. The labeling of target DNA nucleotides in the TCC and STC active sites indicates the utilized oligonucleotide sequence and position of DNA strand transfer (26). Metal ion A in the SSC activates a water molecule to process the CA/AT phosphodiester bond, whereas metal ion B in the TCC activates the resulting CAOH 3′-hydroxyl for strand transfer into target DNA (13).