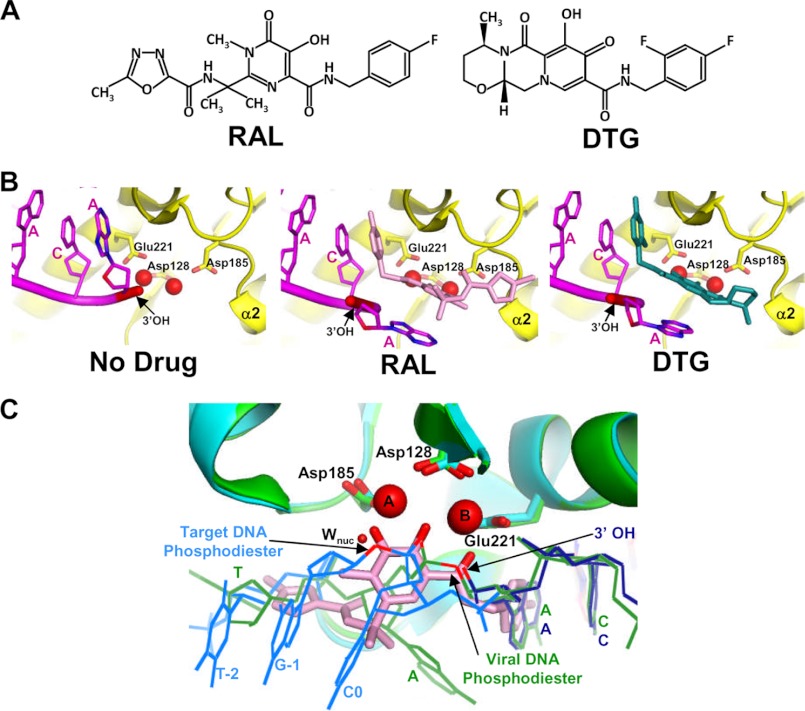
FIGURE 3.
Mechanism of action of INSTIs. A, chemical structures of RAL and DTG. B, comparison of drug-free (left; PDB code 3OY9), RAL-bound (middle; PDB code 3OYA), and DTG-bound (right; PDB code 3S3M) PFV intasomes shows the displacement of the terminal dA nucleotide (carbon, oxygen, and nitrogen atoms are colored magenta, red, and blue, respectively) and the reactive 3′-OH (colored red and marked by arrows) of the transferred DNA strand. C, IN active sites from the SSC (green; PDB code 4E7I), TCC (cyan; PDB code 4E7K), and RAL-bound (PDB code 3OYA; only the drug, in pink sticks, is shown) CDC are superimposed using the Cα atoms of active site residues Asp-128, Asp-185, and Glu-221. The unprocessed viral DNA end in the SSC is green, whereas the processed viral DNA and bound target DNA strands from the TCC structure are in dark blue and cyan, respectively. Mn2+ ions (labeled A and B), metal-chelating oxygens of RAL, bridging oxygen atoms of the scissile viral DNA and target DNA phosphodiester bonds (arrows), and 3′-processing (Wnuc, sphere) and DNA strand transfer (3′-OH, arrow) nucleophiles are colored red to highlight substrate mimicry of INSTIs during retroviral DNA integration (13).