Background: Protein folding mediated by a symmetric GroEL-(GroES)2 complex is poorly understood.
Results: Single-molecule imaging revealed that GFP folding proceeded in both rings of the symmetric GroEL-(GroES)2 complex.
Conclusion: The same reaction occurs independently in both rings of the symmetric GroEL-(GroES)2 complex.
Significance: The study provides insight into the mechanism of protein folding mediated by the symmetric GroEL-(GroES)2 complex.
Keywords: Chaperone Chaperonin, Fluorescence, Microscopy, Protein Folding, Single Molecule Biophysics, GroEL
Abstract
The chaperonin, GroEL, is an essential molecular chaperone that mediates protein folding together with its cofactor, GroES, in Escherichia coli. It is widely believed that the two rings of GroEL alternate between the folding active state coupled to GroES binding during the reaction cycle. In other words, an asymmetric GroEL-GroES complex (the bullet-shaped complex) is formed throughout the cycle, whereas a symmetric GroEL-(GroES)2 complex (the football-shaped complex) is not formed. We have recently shown that the football-shaped complex coexists with the bullet-shaped complex during the reaction cycle. However, how protein folding proceeds in the football-shaped complex remains poorly understood. Here, we used GFP as a substrate to visualize protein folding in the football-shaped complex by single-molecule fluorescence techniques. We directly showed that GFP folding occurs in both rings of the football-shaped complex. Remarkably, the folding was a sequential two-step reaction, and the kinetics were in excellent agreement with those in the bullet-shaped complex. These results demonstrate that the same reactions take place independently in both rings of the football-shaped complex to facilitate protein folding.
Introduction
Chaperonins are a major class of molecular chaperones that assist protein folding in the cell (1, 2). The best characterized chaperonin is Escherichia coli GroEL, which functions together with its cofactor, GroES (3, 4). GroEL is a large, cylindrical complex arranged as two heptameric rings composed of 57-kDa subunits, and these rings are stacked back-to-back. The cochaperonin, GroES, is arranged as a single heptameric ring composed of 10-kDa subunits that interacts with GroEL in a nucleotide-dependent manner.
The commonly accepted model of GroEL-mediated protein folding is as follows. First, one of the GroEL rings captures a denatured protein via its hydrophobic sites, and GroES binds to the same ring in an ATP-dependent manner (the ring is called the cis-ring). GroES binding induces the displacement of the captured protein into the GroEL cavity, where productive folding proceeds for proteins up to ∼60 kDa (5). Next, hydrolysis of ATP in the cis-ring is followed by subsequent ATP binding to the opposite ring (trans-ring), which causes the dissociation of GroES and ADP and release of the substrate protein from the cis-ring. At the same time, the second GroES binds to the trans-ring to reorient a new cis-ring and starts the next ATPase cycle (6, 7). In short, because GroES binds alternatively to each ring of GroEL (two-stroke model), an asymmetric GroEL-GroES complex (termed the bullet-shaped complex) exists throughout the reaction cycle, whereas a symmetric GroEL-(GroES)2 complex (termed the football-shaped complex) is not formed.
In contrast, the existence of the football-shaped complex in the presence of ATP has been suggested by electron microscopic observation (8–10) and biochemical experiments (11–13). In addition, our recent studies have shown that the football-shaped and bullet-shaped complexes coexist during the reaction cycle (14), and the formation of the football-shaped complex is promoted by increases in the ATP/ADP ratio (14) and the concentration of denatured protein (15). We have also described the dynamics of the GroEL-GroES interaction, including the football-shaped complex, in the steady state (16). Several studies have suggested that denatured proteins can be encapsulated and folded in both rings of GroEL at the same time (17–21). However, how protein folding proceeds in the football-shaped complex is unclear.
Here, we used GFP as a substrate to visualize protein folding in the football-shaped complex by single-molecule approaches. We found that two GFP molecules can refold in both rings of the football-shaped complex. We also found that the folding of GFP in each ring of the football-shaped complex is a sequential two-step reaction, and the kinetics are similar to those in the bullet-shaped complex. Our results demonstrate that the same reactions occur in both rings of the football-shaped complex.
EXPERIMENTAL PROCEDURES
Reagents
Apyrase from potato, acetylated BSA, glucose oxidase from Aspergillus niger, catalase from bovine liver, and hexokinase from Saccharomyces cerevisiae were obtained from Sigma-Aldrich. Streptavidin, 1-(2-nitrophenyl)ethyl-caged ATP, and 1-(2-nitrophenyl)ethyl-caged ADP were from Invitrogen. ATP was from Roche Applied Science. Maleimide PEO2-biotin was from Thermo Scientific. Cy5-maleimide and Cy5 monofunctional N-hydroxysuccinimide ester were obtained from GE Healthcare. Other reagents were purchased from Wako Pure Chemicals. Biotinylated BSA was prepared as described (22). ADP was treated with hexokinase in the presence of glucose to eliminate contaminating ATP as described previously (18). Caged ATP and caged ADP were treated with apyrase to remove contaminating ATP and ADP.
Preparation of GroEL and GroES
The GroEL and GroES variants used in this study were as follows: the D398A variant of GroEL (EL398)2 (6), the single-ring variant of EL398 (SR398) (6), the A133C/D398A variant of GroEL (EL133/398) (23), and the GroES variant ES98C, which has a cysteine residue added at the C terminus of each subunit (24). The variants were obtained by site-directed mutagenesis. GroEL, GroES, and their variants were expressed in E. coli cells and purified as previously described (25). Purified proteins were precipitated in 65% saturated ammonium sulfate and stored at 4 °C.
To count the number of GFP molecules, wild-type GroEL (wtEL), EL398, and SR398 were labeled with Cy5 monofunctional N-hydroxysuccinimide ester in HKM buffer (25 mm HEPES-KOH, pH 7.4, 100 mm KCl, and 5 mm MgCl2) containing 20 mm sodium bicarbonate to raise the pH (∼8.5). ES98C was labeled with maleimide PEO2-biotin in HKM buffer. To monitor SBP-GFP folding, EL133/398 was labeled with Cy5-maleimide and maleimide PEO2-biotin. Free label was separated from labeled proteins by gel filtration on NAP5 columns (GE Healthcare). Protein and dye concentrations were determined by absorption spectroscopy with a spectrophotometer (V-550; Jasco), using the following molar extinction coefficients: GroEL tetradecamer, 130,480 m−1 cm−1 at 280 nm; GroEL heptamer, 65,240 m−1 cm−1 at 280 nm; GroES heptamer, 8,960 m−1 cm−1 at 280 nm; and Cy5, 250,000 m−1 cm−1 at 649 nm. The concentrations of Cy5-labeled (and biotin-labeled) GroEL (Cy5-wtEL, Cy5-EL398, Cy5-SR398, and Cy5bio-EL133/398) were determined by correcting for the absorbance of Cy5 at 280 nm. The molar ratios of Cy5 to wtEL, EL398, SR398, and EL133/398 were 0.93, 0.87, 0.79, and 1.2, respectively. Fluorescently labeled or biotinylated GroEL and GroES exhibited behaviors similar to those of the wild-type proteins (data not shown).
Preparation of GFP and SBP-GFP
The GFP employed in this study was the S65T mutant (26). GFP with the strong binding peptide tag (SBP-tag, SWMTTPWGFLHP) at the N terminus (termed SBP-GFP) was obtained using the KOD-Plus-mutagenesis kit (TOYOBO) (27–30). E. coli BL21(DE3) cells transformed with the expression plasmids were grown at 37 °C until the A600 reached 0.5–0.6 and then cultivated in the presence of 1 mm isopropylthio-β-galactoside for 18 h at 15 °C to express GFP and SBP-GFP in the soluble fraction. The recombinant proteins were purified using TOYOPEARL Butyl-650 (Tosoh) and Resource Q (GE Healthcare) columns. The concentrations of GFP and SBP-GFP were determined spectrophotometrically at 280 nm with extinction coefficients of 18,850 and 31,150 m−1 cm−1, respectively.
Counting the Number of GFP Molecules in GroEL-GroES Complexes
GFP (5 μm) was denatured in HKM buffer containing 0.1 m HCl for 5 min at 23 °C. The denatured GFP was diluted to a final concentration of 100 nm in HKM buffer containing 25 nm Cy5-EL398 or 50 nm Cy5-SR398, 500 nm bio-ES98C, and 5 mm dithiothreitol. One minute after the dilution, nucleotide(s) (1 mm ATP or 1 mm ATP + 1 mm ADP) were added. After incubation for 1 min at room temperature, the mixture was diluted, and GroEL-GroES complexes were immobilized on the surface of the coverslips via biotinylated BSA and streptavidin as described (31). Experimental results were obtained in HKM buffer containing nucleotide(s) and an oxygen scavenging system (25 mm glucose, 50 units/ml glucose oxidase, 50 units/ml catalase, and 10 mm dithiothreitol).
Observations were carried out at 23 °C by objective-type total internal reflection fluorescence microscopy (TIRFM) (32). The specimen was illuminated with a 635-nm laser (Radius 635-25; Coherent) to mark the position of Cy5-labeled GroEL and then illuminated with a 488-nm laser (Sapphire 488-75 CDRH; Coherent) to visualize GFP fluorescence through an inverted microscope (IX-71; Olympus) with an oil immersion objective (UApo 150× OTIRF, NA 1.45; Olympus) and dichroic mirrors (a custom-made mirror for Cy5; Chroma Technology and 505DRLP for GFP; Omega Optical). Fluorescence signals from Cy5 and GFP were collected through an objective on a microscope equipped with dichroic mirrors and emission filters (FF01-692/40-25 for Cy5 and FF01-520/35-25 for GFP; Semrock). Fluorescence images were recorded using an electron multiplying charge-coupled device camera (C9100-13; Hamamatsu Photonics) with a time resolution of 200 ms.
Real Time Imaging of SBP-GFP Folding
SBP-GFP (5 μm) was denatured in HKM buffer containing 0.1 m HCl for 5 min at 23 °C. The denatured SBP-GFP was diluted to a final concentration of 100 nm with HKM buffer containing 25 nm Cy5bio-EL133/398. After incubation for 5 min, the mixture was diluted and EL133/398 complexed with denatured SBP-GFP was immobilized on the quartz slide surface via biotinylated BSA and streptavidin as described (31, 33). Experimental results were obtained in HKM buffer containing caged nucleotide(s) (1 mm caged ATP or 1 mm caged ATP + 1 mm caged ADP), 2 μm GroES, and an oxygen scavenging system.
The appearance of SBP-GFP fluorescence at the position of EL133/398 after photolysis of caged ATP was visualized at 23 °C by prism-type TIRFM (33, 34). The specimen was illuminated with a 635-nm laser to mark the position of Cy5-EL133/398 and then illuminated with a 488-nm laser to visualize SBP-GFP fluorescence. Fluorescence signals from Cy5 and SBP-GFP were collected through an objective (UApo 150× OTIRF) on a microscope (IX-71) equipped with a dichroic mirror (FF409-Di03-25×36, Semrock), a long pass filter (FF01-409/LP-25, Semrock) and emission filters (FF01-692/40-25 and FF01-520/35-25). Fluorescence images were obtained with an electron multiplying charge-coupled device camera with a time resolution of 200 ms. For photolysis of caged nucleotides, the specimens were illuminated with a 250-ms pulse of UV light from a 100-W mercury lamp (USH-102D; Ushio Lighting) via a band-pass filter (FF01-357/44-25; Semrock) and a dichroic mirror (FF409-Di03-25×36). Approximately 35% of the caged nucleotides were split under the experimental conditions.
Data Analysis
The recorded images were analyzed using a homemade program on a Halcon image processor (MVTec Software GmbH, Munich, Germany) to obtain time trajectories of the fluorescence intensities of Cy5 and GFP molecules. The number of folded SBP-GFP molecules was plotted as a function of time. In the football condition, the number of molecules is expressed as the sum of the number of the first refolded and second refolded SBP-GFP molecules. The histograms of the time required for SBP-GFP folding were fitted by using the following equation,
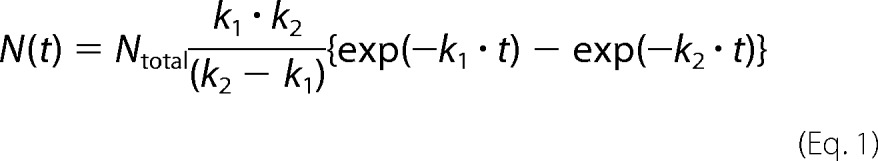 |
where N(t) is the number of refolded SBP-GFP molecules in GroEL-GroES complexes at time t and Ntotal is the total number of refolded SBP-GFP molecules. In addition, k1 and k2 are the rate constants of a two-step reaction with different rate constants. Rate constants for SBP-GFP folding were determined by fitting to the cumulative frequency distributions using the following equation.
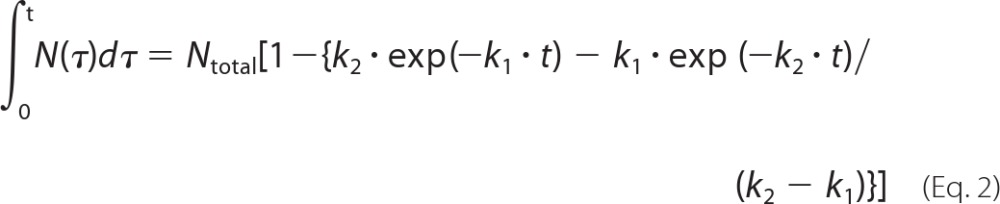 |
Data fitting was performed using the Kaleidagraph program (Synergy Software).
RESULTS
Protein Folding Can Proceed in Each Ring of the Football-shaped GroEL-GroES Complex
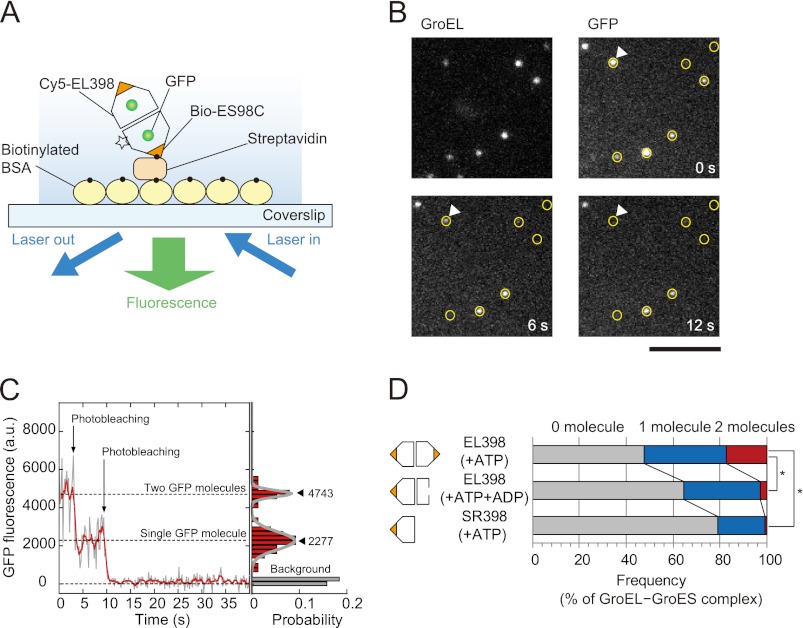
First, we examined how many GFP molecules can fold within the football-shaped complex at the single-molecule level. To this end, the D398A variant of GroEL (EL398) was used. EL398 hydrolyzes ATP much more slowly than wtEL and then forms a long-lived football-shaped complex upon the addition of ATP to retain refolded GFP molecules in the complex (half-time of ∼30 min) (6, 14, 21). EL398 was fluorescently labeled with Cy5 monofunctional N-hydroxysuccinimide ester (termed Cy5-EL398) because it does not contain reactive cysteine residues. A cysteine-introduced GroES variant (ES98C) (24) was modified with biotin-maleimide (termed bio-ES98C). Cy5-EL398 (25 nm) and bio-ES98C (500 nm) were incubated with acid-denatured GFP (100 nm) and nucleotide(s) to form GroEL-GroES complexes. The complexes were immobilized on a glass surface via a biotin-streptavidin linkage (Fig. 1A). Observations of single-molecule fluorescence were performed by objective-type TIRFM. The positions of the GroEL-GroES complexes were determined from the fluorescence of Cy5 attached to EL398 (Fig. 1B, GroEL). As expected, the fluorescence of GFP molecules was detected at the positions of the GroEL-GroES complexes (Fig. 1B, GFP), indicating that refolded GFP was encapsulated in the GroEL-GroES complexes. In the fluorescence time traces of GFP, a stepwise decrease in fluorescence intensity was observed, which reflected the photobleaching of individual GFP molecules (Fig. 1C). The number of photobleaching steps was equivalent to the number of refolded GFP molecules within the GroEL-GroES complexes. Then we counted the number of steps required for photobleaching of the GFP molecules to determine the number of GFP molecules within the GroEL-GroES complexes (Fig. 1D).
FIGURE 1.
Number of GFP molecules in GroEL-GroES complexes. A, schematic illustration of the experiment. The GroEL-GroES complexes formed in the presence of denatured GFP were immobilized on the glass surface of the coverslips via biotinylated BSA and streptavidin. The flow cell was filled with HKM buffer containing nucleotide(s) and an oxygen scavenging system. The fluorescence of Cy5 and GFP was observed by objective-type TIRFM. The number of GFP molecules in the GroEL-GroES complex was determined by counting the number of photobleaching steps. The experimental details are described under “Experimental Procedures.” B, fluorescence images of Cy5-EL398 and GFP molecules. The positions of Cy5-EL398 are indicated by circles on the fluorescence images of GFP. GFP fluorescence that did not appear at the position of marked Cy5-EL398 may be attributable to nonlabeled EL398. The scale bar represents 5 μm. C, left panel, time trajectory of the fluorescence intensity of GFP at the position indicated by white triangles in Fig. 1B. Raw data (gray) are overlaid with averaged traces (red, 1 s-moving average). The trajectory shows the two-step photobleaching events of GFP molecules, indicating that two refolded GFP molecules are encapsulated in the GroEL-GroES complex. Right panel, histogram of the fluorescence intensity of GFP for 15 s after illumination. The histogram was fitted by the sum of two Gaussian functions. Closed triangles represent the peak of the Gaussian (the mean fluorescence intensity). D, number of GFP molecules in football-shaped and bullet-shaped complexes. Each value represents the average of three independent experiments (n = 192–228 in one experiment). To assess the differences between the percentages of the GroEL-GroES complexes containing two GFP molecules, statistical analysis was performed using Student's t test. The asterisk indicates p < 0.01.
In the presence of ATP, conditions in which EL398 forms the football-shaped complexes (14), two GFP molecules were observed in 17 ± 1.2% (mean ± S.E.) of the GroEL-GroES complexes, whereas one GFP molecule was observed in 35 ± 2.5% (mean ± S.E.) of the complexes (Fig. 1D, row 1). Three GFP molecules were rarely detected in the complexes (less than 0.3%). Only half of GroEL-GroES complexes contain refolded GFP because denatured GFP can be easily released from GroEL-GroES complexes into the bulk medium (35). Our data indicated that two GFP molecules can refold in the football-shaped complex. However, there remained the possibility that two GFP molecules refolded in the same ring, not in both rings. In an earlier study (5), it was shown that a fusion protein of GFP and its blue fluorescent variant can fold in the cis-ring. To exclude the possibility that two GFP molecules fold in one ring of the GroEL-GroES complexes, control experiments were performed. In the presence of both ATP and ADP, EL398 forms the bullet-shaped complexes because ADP inhibits the binding of ATP to the trans-ring of GroEL (14). With the addition of ATP and ADP to EL398, the percentage of the GroEL-GroES complexes containing two GFP molecules was drastically reduced to 2.7 ± 0.70% (mean ± S.E.; Fig. 1D, row 2), indicating that one GFP molecule completed the folding in the bullet-shaped complex. We also used the single-ring variant of EL398 (SR398) (6). The detection rate of two GFP molecules in SR398-GroES complexes was negligible (0.78 ± 0.13%, mean ± S.E.; Fig. 1D, row 3). These experiments demonstrated that two GFP molecules did not refold in the same ring under the experimental conditions used. In summary, GFP can be encapsulated and refold in both rings of the football-shaped complex.
SBP-GFP Refolds with the Same Kinetics in Both Rings of the Football-shaped GroEL-GroES Complex
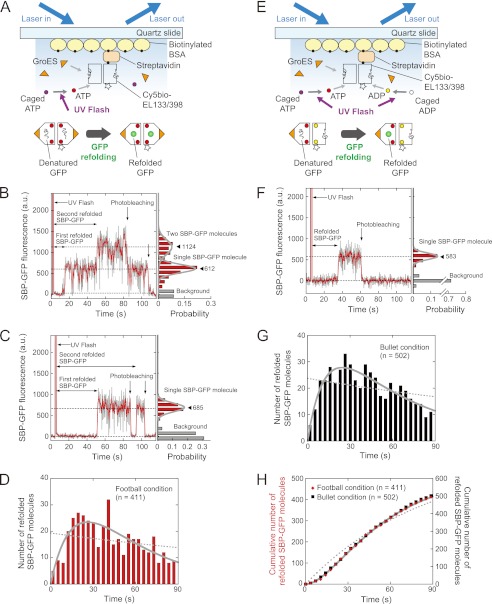
We next performed real time imaging of GFP folding in the football-shaped and bullet-shaped complexes (Fig. 2, A and E). To this end, we prepared an EL398 variant in which Ala-133 located on the outer surface of the equatorial domain was mutated to cysteine (EL133/398) (23). EL133 was easily and specifically modified with fluorophore and biotin, and the chaperonin function was not perturbed on the glass surface (data not shown). EL133/398 was modified with Cy5-maleimide and biotin-maleimide (termed Cy5bio-EL133/398). Cy5bio-EL133/398 complexed with acid-denatured GFP was immobilized on the quartz slide via a biotin-streptavidin linkage to allow the folding of GFP to be monitored. The final solution contained 2 μm GroES and caged nucleotide(s) (1 mm caged ATP or 1 mm caged ATP + 1 mm caged ADP), and an oxygen scavenging system. After confirming the positions of the Cy5bio-EL133/398 molecules, a UV flash was used to illuminate the specimen for the photogeneration of nucleotide(s) while monitoring the fluorescence from GFP molecules within the GroEL-GroES complexes by prism-type TIRFM (Fig. 2, A and E). Unexpectedly, refolding of two GFP molecules was observed in only 3% of the complexes in these conditions. This could be caused by the irreversible dissociation of denatured GFP from GroEL during the preparation process before the UV flash (∼20 min). To solve this problem, we employed a GFP variant in which SBP (SWMTTPWGFLHP) was fused to the N terminus (termed SBP-GFP). SBP binds to the substrate recognition site of GroEL with high affinity (27), and the peptide and denatured protein fused to the SBP-tag bind to GroEL with higher affinity (28–30). In fact, by using SBP-GFP, the percentage of the GroEL-GroES complexes containing two GFP molecules was significantly increased to 13%.
FIGURE 2.
Real time imaging of SBP-GFP folding inside GroEL-GroES complexes. A, schematic illustration of the experiment for SBP-GFP refolding in the football-shaped complex. Cy5-EL133/398 complexed with denatured GFP was immobilized on the quartz slide surface via biotinylated BSA and streptavidin. The final solution contained saturating amounts of GroES and caged ATP, and an oxygen scavenging system. After the photolysis of caged ATP, denatured GFP molecules initiated the folding inside the football-shaped complex. Fluorescence signals from Cy5 and GFP were obtained by prism-type TIRFM. B and C, left panels, time trajectories for the fluorescence intensity of SBP-GFP molecules under the football condition. The second folded SBP-GFP molecule appeared before (B) or after (C) the photobleaching of the first folded SBP-GFP molecule. B and C, right panel, histograms of the fluorescence intensity of SBP-GFP after photolysis. The histograms were fitted by the sum of two Gaussian functions (B) and a Gaussian function (C), respectively. Closed triangles represent the peak of the Gaussian (the mean fluorescence intensity). D, histogram of the time required for the appearance of SBP-GFP fluorescence in the football-shaped complex. The number of molecules is expressed as the sum of the number of the first refolded and second refolded SBP-GFP molecules. The dotted line was fitted to the data by a single exponential function. The solid line was fitted to the data by using Equation 1. E, schematic illustration of the experiment for SBP-GFP refolding in the bullet-shaped complex. The experimental conditions were similar, except that caged ATP and caged ADP were added to the solution. F, left panel, time trajectory for the fluorescence intensity of SBP-GFP molecules under the bullet condition. The fluorescence of single SBP-GFP molecules was observed under the bullet condition. Right panel, histogram of the fluorescence intensity of SBP-GFP after photolysis. The histogram was fitted by using a Gaussian function. G, histogram of the time required for the appearance of SBP-GFP fluorescence in the bullet-shaped complex. The dotted line was fitted to the data by a single exponential function. The solid line was fitted to the data by using Equation 1. H, plots of the cumulative number of refolded SBP-GFP molecules in the football-shaped complex (red diamonds), and the bullet-shaped complex (black squares). In the football condition, the number of molecules is expressed as the sum of the number of the first refolded and second refolded SBP-GFP molecules. The dotted and solid lines represent the fitting curve for the football condition data. The dotted line was fitted to the data by a single exponential function. The solid line was fitted to the data using Equation 2.
Football-shaped complexes were formed by the photogeneration of ATP (termed the football condition). In the football condition, the appearance of two fluorescent species was observed at the position of EL133/398, reflecting that two SBP-GFP molecules were refolded in the football-shaped complex (Fig. 2, B and C). As evidence of this, the observed fluorescence blinking of SBP-GFP in the football-shaped complex occurred only infrequently (∼2%) under these experimental conditions. The second folded SBP-GFP molecules appeared before and after the photobleaching of the first folded SBP-GFP molecules (Fig. 2, B and C). In contrast, bullet-shaped complexes were formed by the photogeneration of ATP and ADP from caged ATP and caged ADP (termed the bullet condition). In the bullet condition, a single fluorescent molecule appeared at the position of EL133/398, indicating that one SBP-GFP molecule was refolded in the bullet-shaped complex (Fig. 2F).
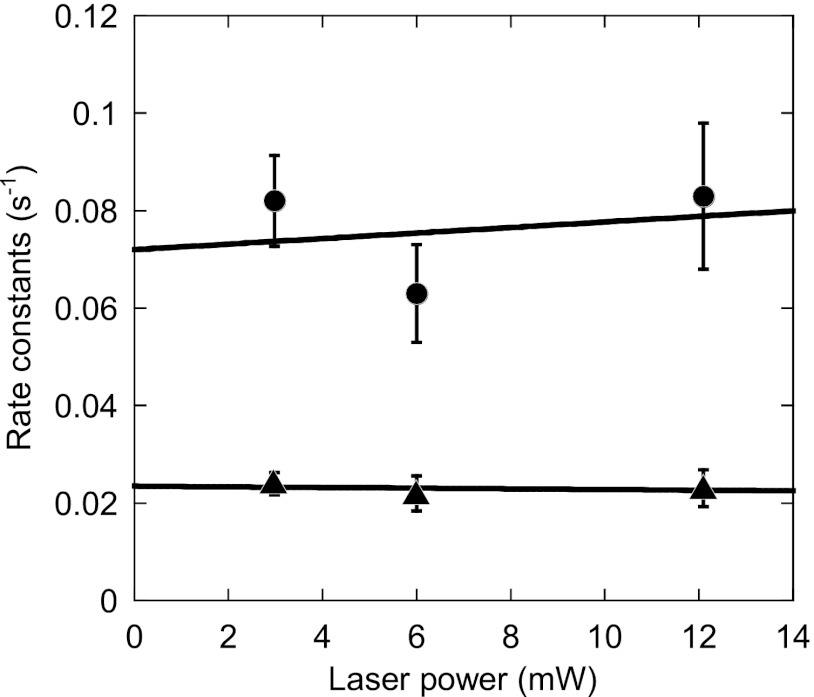
We analyzed the time until the appearance of SBP-GFP fluorescence after the photolysis of caged nucleotides in the football and bullet conditions and constructed the histograms (Fig. 2, D and G). In the football condition (Fig. 2A), the number of molecules is expressed as the sum of the number of the first refolded and second refolded SBP-GFP molecules. Both histograms have maximum peaks at ∼25 s. The histograms are not fitted by a single exponential function (χ2 = 66, p < 0.05 in the football condition; χ2 = 47, p < 0.05 in the bullet condition) (Fig. 2, D and G, dotted lines). On the other hand, the histograms are well fitted by Equation 1, which is derived from the sequential two-step reaction with different rate constants (χ2 = 25, p > 0.1 in the football condition; χ2 = 11, p > 0.1 in the bullet condition) (Fig. 2, D and G, solid lines). These results indicate that the folding of SBP-GFP is not governed by a single rate-limiting step but rather obeys two sequential steps, as shown in our previous study (33). We then plotted the time courses for the accumulation of fluorescent SBP-GFP molecules to precisely determine the rate constants (Fig. 2H). The plots are well fitted by Equation 2, which is obtained by integration of Equation 1 over time (solid line). Note that the order of the reactions governed by the rate constants k1 and k2 cannot be determined mathematically. The faster rate constants in the football-shaped and bullet-shaped complexes were determined to be 0.063 ± 0.010 and 0.065 ± 0.0077 s−1, respectively (values are reported with the errors of the fits). The slower rate constants in the football-shaped and bullet-shaped complexes were 0.022 ± 0.0036 and 0.021 ± 0.0025 s−1, respectively. These results indicate that SBP-GFP folding in the football-shaped complex proceeded with the same kinetics as in the bullet-shaped complex. We also determined the rate constants of SBP-GFP folding in the football-shaped complex at the different excitation laser power conditions (Fig. 3). The rate constants obtained are independent of excitation laser power, indicating that the photophysical properties of GFP have little influence on determination of the rate constants.
FIGURE 3.
Rate constants of SBP-GFP folding in the football-shaped complex as a function of laser power. Real time imaging of SBP-GFP folding in the football-shaped complex was performed as described under “Experimental Procedures” and in Fig. 2 with different laser power conditions. The rate constants of SBP-GFP folding were obtained by fitting the data by using Equation 2. The circles indicate the faster rate constants, and triangles indicate the slower rate constants. The values are reported with the errors of the fits. The solid lines were obtained by the least square method.
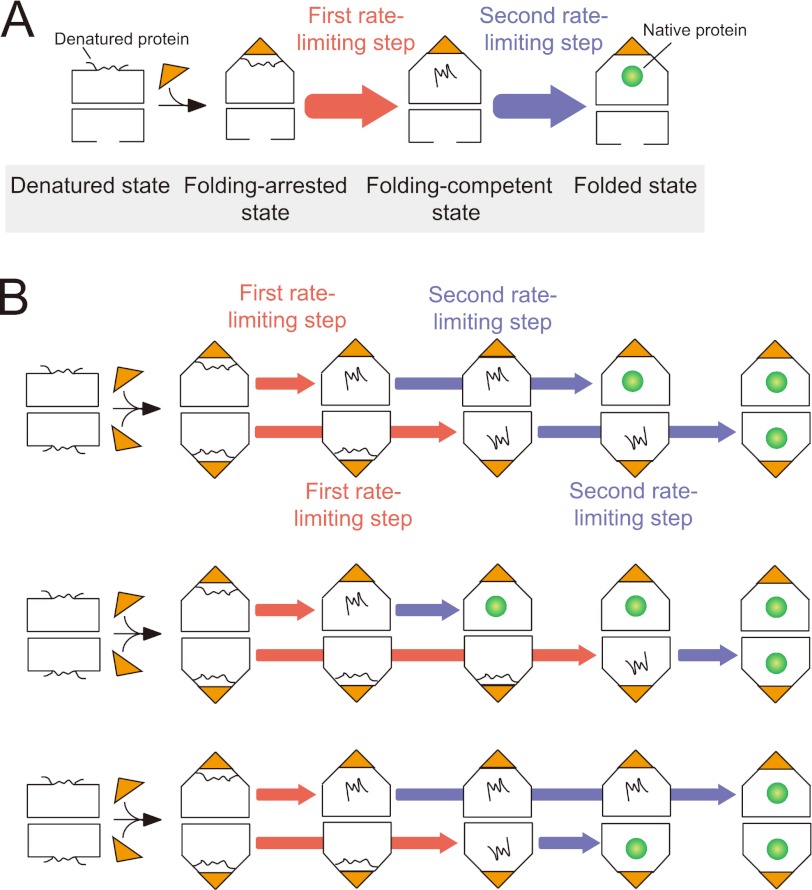
We have previously shown that protein folding occurs in a two-step reaction in the bullet-shaped complex (33, 36) (Fig. 4A). The proposed model will be applicable to protein folding in the football-shaped complex (Fig. 4B). As indicated previously, in the first rate-limiting step, the folding of the substrate proteins is arrested in the GroEL-GroES cavity (folding-arrested state). In the second rate-limiting step, the substrate is released into the central cavity (folding-competent state), where productive folding occurs (folded state). Given that the slower step (k = 0.022 s−1) proceeds with roughly the same kinetics as the spontaneous folding of SBP-GFP in bulk measurement (k = 0.035 s−1), the rate constants of the first and second rate-limiting steps can be estimated to be 0.063 and 0.022 s−1, respectively. These results indicated that two GFP molecules can independently refold with two rate-limiting steps in both rings of the football-shaped complex.
FIGURE 4.
Schematic model of protein folding in the bullet-shaped and football-shaped GroEL-GroES complexes. A, protein folding in the bullet-shaped complex. B, protein folding in the football-shaped complex. In the first rate-limiting step (red), the folding of substrate proteins is arrested (folding-arrested state). In the second rate-limiting step (blue), the substrate proteins are released into the cavity (folding-competent state), and protein folding occurs (folded state). The same reactions occur independently in both rings of the football-shaped complex. The upper and lower rings can be inverted. Turnover of ATP hydrolysis is not included in this model.
DISCUSSION
In this study, we used GFP as a substrate to visualize protein folding in the football-shaped GroEL-(GroES)2 complex by single-molecule fluorescence techniques. In general, fluorescence correlation spectroscopy is a powerful method for studying protein folding dynamics in solution (37). However, GFP folding events within the same GroEL-GroES complex over time could not be monitored, because fluorescence correlation spectroscopy monitors some refolded GFP going in and out the focal spot by Brownian motion. Additionally, lipid vesicles can serve as an ideal container for single-molecule fluorescence measurements of protein folding (38). It should be noted that some populations of denatured GFP molecules escape from GroEL and refold spontaneously (35). Therefore, it is technically very difficult to determine the number of refolded GFP molecules and to monitor the folding of GFP only within the GroEL-GroES complexes. Taken all together, we think that surface immobilization is the best strategy employed.
First, we determined how many GFP molecules can fold within the football-shaped complex at the single-molecule level. Previous studies have shown that ∼2 mol of rhodanese/mol of GroEL tetradecamer were present in the football-shaped complex by densitometry analysis and enzymatic activity assay (19, 21). In this study, the number of fluorescent GFP molecules within the football-shaped complex was determined by counting the number of photobleaching steps. We directly observed that single GFP molecules are encapsulated and fold in both rings of the football-shaped complex. Our results clearly demonstrate that protein folding can occur in both rings of the football-shaped complex.
We next performed real time imaging of GFP folding in the football-shaped and bullet-shaped complexes. These experiments showed that the folding of SBP-GFP in the football-shaped complex is a sequential two-step reaction with rate constants of 0.063 and 0.022 s−1. It should be noted that the kinetics in the football-shaped complex were in excellent agreement with those in the bullet-shaped complex, demonstrating that both rings of the football-shaped complex can work independently as a cis-ring in the bullet shaped complex. We have previously investigated the decay process of the football-shaped complex to the bullet-shaped complex at the single-molecule level. We found that the first GroES that interacts with GroEL does not always dissociate from the football-shaped complex prior to the dissociation of the second GroES molecule, i.e., dissociation of GroES molecules from the football-shaped complex can occur in a random order (16). Taken together, there seems to be a lack of inter-ring communication in the football-shaped complex. It has long been thought that ATP cannot bind to the trans-ring until ATP is hydrolyzed in the cis-ring, because inter-ring negative cooperativity (39). Recently, it has been shown that ADP remains in the GroEL ring even after GroES has detached from the same ring (40, 41). Also, it has been demonstrated that ADP prevents the association of ATP (42) and the second GroES with the trans-ring (13, 14). As indicated in our previous report (15), the binding of the denatured protein to the trans-ring will facilitate the dissociation of ADP from the trans-ring and the formation of the football-shaped complex. As a result, the cis-ring and the trans-ring cannot be distinguished. The previously published model of inter-ring negative cooperativity is correct but incomplete in the sense that it does not apply to conditions in which the concentration of denatured protein is high.
SBP-GFP refolded in the GroEL-GroES complex in a two-step reaction with rate constants of 0.063 and 0.022 s−1. In contrast, when GFP was used as a substrate protein, the rate constants for the first and second rate-limiting steps were 0.34 ± 0.077 and 0.041 ± 0.0036 s−1, respectively. It is possible that SBP interacts with not only the apical domain of GroEL but also the inner wall of the GroEL-GroES cavity. However, this has no significant effect on the main conclusion that the same reaction occurs in both rings of the football-shaped complex to facilitate protein folding.
We have found the existence of two cycles in the GroEL-GroES interaction: the bullet cycle and the football cycle (15, 16). At a low concentration of denatured protein, GroEL mainly goes through the bullet cycle because of the inhibitory effect of ADP in the trans-ring. In contrast, in the presence of a high concentration of denatured protein, GroEL works in a different mode to switch to the football cycle. Our data do not contradict the published model, but rather show that the GroEL-GroES system can work in a different mode at a high substrate concentration.
The ATPase activity of GroEL is higher when the levels of the football-shaped complex increase (15). GroEL may display inter-ring negative cooperativity at relatively low substrate concentration to minimize ATP consumption. The football-shaped complex is expected to be a more productive intermediate than the bullet-shaped complex for preventing the accumulation of denatured protein, particularly in the presence of increasing amounts of denatured protein. This mechanism will have enormous significance during the accumulation of stress-induced denatured proteins. As we have shown, GroEL can increase the protein folding capacity by forming the football-shaped complex without waiting for the expression of chaperone proteins. This is one reason why GroEL functions as a double-ring complex. The physiological significance of the football-shaped complex in vivo remains to be seen.
Acknowledgments
We thank Dr. Tatsuya Nojima (Frontier Research Center, Tokyo Institute of Technology) for helpful advice on SBP-GFP, and Dr. Yoshitaka Shirasaki (Laboratory for Immunogenomics, Research Center for Allergy and Immunology, RIKEN) for image analysis programming.
This work was partly supported by Grants-in-Aid for Scientific Research (B) 24370061 (to T. F.), Scientific Research on Innovative Areas 21121004 (to T. F.), and Scientific Research on Priority Areas 22020006 (to R. I.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This research was also supported by the Funding Program for World-Leading Innovative R & D on Science and Technology (FIRST Program) from the Japan Society for the Promotion of Science.
- EL398
- the D398A variant of GroEL
- wtEL
- wild-type GroEL
- SR398
- the single-ring variant of EL398
- EL133/398
- the A133C/D398A variant of GroEL
- Cy5-EL398
- Cy5-labeled EL398
- Cy5-SR398
- Cy5-labeled SR398
- Cy5bio-EL133/398
- Cy5- and biotin-labeled EL133/398
- ES98C
- GroES mutant with a cysteine residue added at the C-terminus of each subunit
- bio-ES98C
- biotin-labeled ES98C
- SBP
- strong binding peptide
- SBP-GFP
- the GFP variant with SBP-tag at the N-terminus
- TIRFM
- total internal reflection fluorescence microscopy.
REFERENCES
- 1. Horwich A. L., Fenton W. A., Chapman E., Farr G. W. (2007) Two families of chaperonin. Physiology and mechanism. Annu. Rev. Cell Dev. Biol. 23, 115–145 [DOI] [PubMed] [Google Scholar]
- 2. Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 [DOI] [PubMed] [Google Scholar]
- 3. Horwich A. L., Farr G. W., Fenton W. A. (2006) GroEL-GroES-mediated protein folding. Chem. Rev. 106, 1917–1930 [DOI] [PubMed] [Google Scholar]
- 4. Horwich A. L., Fenton W. A. (2009) Chaperonin-mediated protein folding. Using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys. 42, 83–116 [DOI] [PubMed] [Google Scholar]
- 5. Sakikawa C., Taguchi H., Makino Y., Yoshida M. (1999) On the maximum size of proteins to stay and fold in the cavity of GroEL underneath GroES. J. Biol. Chem. 274, 21251–21256 [DOI] [PubMed] [Google Scholar]
- 6. Rye H. S., Burston S. G., Fenton W. A., Beechem J. M., Xu Z., Sigler P. B., Horwich A. L. (1997) Distinct actions of cis and trans ATP within the double ring of the chaperonin GroEL. Nature 388, 792–798 [DOI] [PubMed] [Google Scholar]
- 7. Rye H. S., Roseman A. M., Chen S., Furtak K., Fenton W. A., Saibil H. R., Horwich A. L. (1999) GroEL-GroES cycling. ATP and nonnative polypeptide direct alternation of folding-active rings. Cell 97, 325–338 [DOI] [PubMed] [Google Scholar]
- 8. Schmidt M., Rutkat K., Rachel R., Pfeifer G., Jaenicke R., Viitanen P., Lorimer G., Buchner J. (1994) Symmetric complexes of GroE chaperonins as part of the functional cycle. Science 265, 656–659 [DOI] [PubMed] [Google Scholar]
- 9. Beissinger M., Rutkat K., Buchner J. (1999) Catalysis, commitment and encapsulation during GroE-mediated folding. J. Mol. Biol. 289, 1075–1092 [DOI] [PubMed] [Google Scholar]
- 10. Llorca O., Carrascosa J. L., Valpuesta J. M. (1996) Biochemical characterization of symmetric GroEL-GroES complexes. Evidence for a role in protein folding. J. Biol. Chem. 271, 68–76 [DOI] [PubMed] [Google Scholar]
- 11. Azem A., Kessel M., Goloubinoff P. (1994) Characterization of a functional GroEL14(GroES7)2 chaperonin hetero-oligomer. Science 265, 653–656 [DOI] [PubMed] [Google Scholar]
- 12. Azem A., Diamant S., Kessel M., Weiss C., Goloubinoff P. (1995) The protein-folding activity of chaperonins correlates with the symmetric GroEL14(GroES7)2 heterooligomer. Proc. Natl. Acad. Sci. U.S.A. 92, 12021–12025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nojima T., Yoshida M. (2009) Probing open conformation of GroEL rings by cross-linking reveals single and double open ring structures of GroEL in ADP and ATP. J. Biol. Chem. 284, 22834–22839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sameshima T., Ueno T., Iizuka R., Ishii N., Terada N., Okabe K., Funatsu T. (2008) Football- and bullet-shaped GroEL-GroES complexes coexist during the reaction cycle. J. Biol. Chem. 283, 23765–23773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sameshima T., Iizuka R., Ueno T., Funatsu T. (2010) Denatured proteins facilitate the formation of the football-shaped GroEL-(GroES)2 complex. Biochem. J. 427, 247–254 [DOI] [PubMed] [Google Scholar]
- 16. Sameshima T., Iizuka R., Ueno T., Wada J., Aoki M., Shimamoto N., Ohdomari I., Tanii T., Funatsu T. (2010) Single-molecule study on the decay process of the football-shaped GroEL-GroES complex using zero-mode waveguides. J. Biol. Chem. 285, 23159–231564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sparrer H., Rutkat K., Buchner J. (1997) Catalysis of protein folding by symmetric chaperone complexes. Proc. Natl. Acad. Sci. U.S.A. 94, 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Motojima F., Yoshida M. (2003) Discrimination of ATP, ADP, and AMPPNP by chaperonin GroEL. Hexokinase treatment revealed the exclusive role of ATP. J. Biol. Chem. 278, 26648–26654 [DOI] [PubMed] [Google Scholar]
- 19. Taguchi H., Tsukuda K., Motojima F., Koike-Takeshita A., Yoshida M. (2004) BeFx stops the chaperonin cycle of GroEL-GroES and generates a complex with double folding chambers. J. Biol. Chem. 279, 45737–45743 [DOI] [PubMed] [Google Scholar]
- 20. Koike-Takeshita A., Shimamura T., Yokoyama K., Yoshida M., Taguchi H. (2006) Leu309 plays a critical role in the encapsulation of substrate protein into the internal cavity of GroEL. J. Biol. Chem. 281, 962–967 [DOI] [PubMed] [Google Scholar]
- 21. Koike-Takeshita A., Yoshida M., Taguchi H. (2008) Revisiting the GroEL-GroES reaction cycle via the symmetric intermediate implied by novel aspects of the GroEL(D398A) mutant. J. Biol. Chem. 283, 23774–23781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Itakura S., Yamakawa H., Toyoshima Y. Y., Ishijima A., Kojima T., Harada Y., Yanagida T., Wakabayashi T., Sutoh K. (1993) Force-generating domain of myosin motor. Biochem. Biophys. Res. Commun. 196, 1504–1510 [DOI] [PubMed] [Google Scholar]
- 23. Yamasaki R., Hoshino M., Wazawa T., Ishii Y., Yanagida T., Kawata Y., Higurashi T., Sakai K., Nagai J., Goto Y. (1999) Single molecular observation of the interaction of GroEL with substrate proteins. J. Mol. Biol. 292, 965–972 [DOI] [PubMed] [Google Scholar]
- 24. Murai N., Makino Y., Yoshida M. (1996) GroEL locked in a closed conformation by an interdomain cross-link can bind ATP and polypeptide but cannot process further reaction steps. J. Biol. Chem. 271, 28229–28234 [DOI] [PubMed] [Google Scholar]
- 25. Motojima F., Makio T., Aoki K., Makino Y., Kuwajima K., Yoshida M. (2000) Hydrophilic residues at the apical domain of GroEL contribute to GroES binding but attenuate polypeptide binding. Biochem. Biophys. Res. Commun. 267, 842–849 [DOI] [PubMed] [Google Scholar]
- 26. Heim R., Cubitt A., B., Tsien R., Y. (1995) Improved green fluorescence. Nature 373, 663–664 [DOI] [PubMed] [Google Scholar]
- 27. Chen L., Sigler P. B. (1999) The crystal structure of a GroEL/peptide complex. Plasticity as a basis for substrate diversity. Cell 99, 757–768 [DOI] [PubMed] [Google Scholar]
- 28. Li Y., Gao X., Chen L. (2009) GroEL recognizes an amphipathic helix and binds to the hydrophobic side. J. Biol. Chem. 284, 4324–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tanaka S., Kawata Y., Otting G., Dixon N. E., Matsuzaki K., Hoshino M. (2010) Chaperonin-encapsulation of proteins for NMR. Biochim. Biophys. Acta 1804, 866–871 [DOI] [PubMed] [Google Scholar]
- 30. Nojima T., Ikegami T., Taguchi H., Yoshida M. (2012) Flexibility of GroES mobile loop is required for efficient chaperonin function. J. Mol. Biol. 422, 291–299 [DOI] [PubMed] [Google Scholar]
- 31. Taguchi H., Ueno T., Tadakuma H., Yoshida M., Funatsu T. (2001) Single-molecule observation of protein-protein interactions in the chaperonin system. Nat. Biotechnol. 19, 861–865 [DOI] [PubMed] [Google Scholar]
- 32. Tokunaga M., Kitamura K., Saito K., Iwane A. H., Yanagida T. (1997) Single molecule imaging of fluorophores and enzymatic reactions achieved by objective-type total internal reflection fluorescence microscopy. Biochem. Biophys. Res. Commun. 235, 47–53 [DOI] [PubMed] [Google Scholar]
- 33. Ueno T., Taguchi H., Tadakuma H., Yoshida M., Funatsu T. (2004) GroEL mediates protein folding with a two successive timer mechanism. Mol. Cell 14, 423–434 [DOI] [PubMed] [Google Scholar]
- 34. Funatsu T., Harada Y., Tokunaga M., Saito K., Yanagida T. (1995) Imaging of single fluorescent molecules and individual ATP turnovers by single myosin molecules in aqueous solution. Nature 374, 555–559 [DOI] [PubMed] [Google Scholar]
- 35. Motojima F., Yoshida M. (2010) Polypeptide in the chaperonin cage partly protrudes out and then folds inside or escapes outside. EMBO J. 29, 4008–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Suzuki M., Ueno T., Iizuka R., Miura T., Zako T., Akahori R., Miyake T., Shimamoto N., Aoki M., Tanii T., Ohdomari I., Funatsu T. (2008) Effect of the C-terminal truncation on the functional cycle of chaperonin GroEL. Implication that the C-terminal region facilitates the transition from the folding-arrested to the folding-competent state. J. Biol. Chem. 283, 23931–23939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neuweiler H., Johnson C. M., Fersht A. R. (2009) Direct observation of ultrafast folding and denatured state dynamics in single protein molecules. Proc. Natl. Acad. Sci. U.S.A. 106, 18569–18574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rhoades E., Gussakovsky E., Haran G. (2003) Watching proteins fold one molecule at a time. Proc. Natl. Acad. Sci. U.S.A. 100, 3197–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Horovitz A., Willison K. R. (2005) Allosteric regulation of chaperonins. Curr. Opin. Struct. Biol. 15, 646–651 [DOI] [PubMed] [Google Scholar]
- 40. Grason J. P., Gresham J. S., Widjaja L., Wehri S. C., Lorimer G. H. (2008) Setting the chaperonin timer. The effects of K+ and substrate protein on ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 105, 17334–17338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Madan D., Lin Z., Rye H. S. (2008) Triggering protein folding within the GroEL-GroES complex. J. Biol. Chem. 283, 32003–32013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cliff M. J., Kad N. M., Hay N., Lund P. A., Webb M. R., Burston S. G., Clarke A. R. (1999) A kinetic analysis of the nucleotide-induced allosteric transitions of GroEL. J. Mol. Biol. 293, 667–684 [DOI] [PubMed] [Google Scholar]