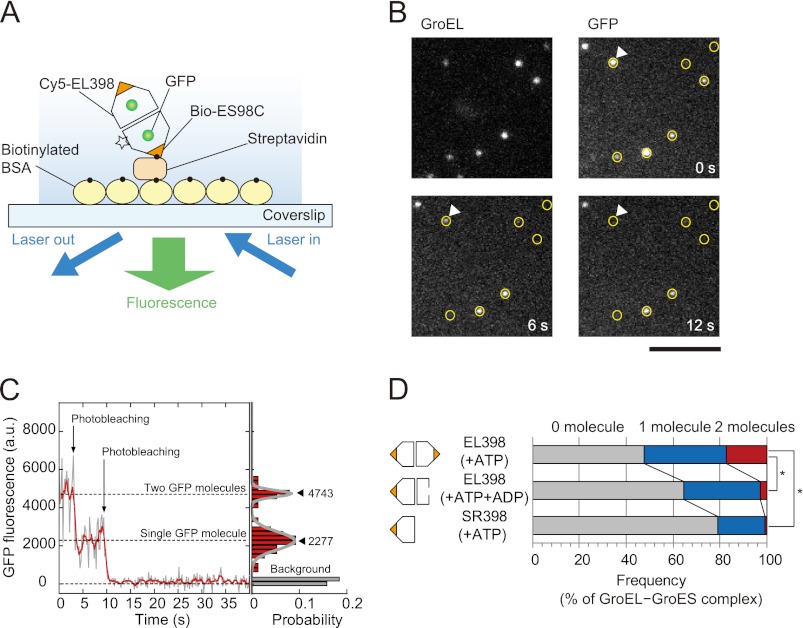
FIGURE 1.
Number of GFP molecules in GroEL-GroES complexes. A, schematic illustration of the experiment. The GroEL-GroES complexes formed in the presence of denatured GFP were immobilized on the glass surface of the coverslips via biotinylated BSA and streptavidin. The flow cell was filled with HKM buffer containing nucleotide(s) and an oxygen scavenging system. The fluorescence of Cy5 and GFP was observed by objective-type TIRFM. The number of GFP molecules in the GroEL-GroES complex was determined by counting the number of photobleaching steps. The experimental details are described under “Experimental Procedures.” B, fluorescence images of Cy5-EL398 and GFP molecules. The positions of Cy5-EL398 are indicated by circles on the fluorescence images of GFP. GFP fluorescence that did not appear at the position of marked Cy5-EL398 may be attributable to nonlabeled EL398. The scale bar represents 5 μm. C, left panel, time trajectory of the fluorescence intensity of GFP at the position indicated by white triangles in Fig. 1B. Raw data (gray) are overlaid with averaged traces (red, 1 s-moving average). The trajectory shows the two-step photobleaching events of GFP molecules, indicating that two refolded GFP molecules are encapsulated in the GroEL-GroES complex. Right panel, histogram of the fluorescence intensity of GFP for 15 s after illumination. The histogram was fitted by the sum of two Gaussian functions. Closed triangles represent the peak of the Gaussian (the mean fluorescence intensity). D, number of GFP molecules in football-shaped and bullet-shaped complexes. Each value represents the average of three independent experiments (n = 192–228 in one experiment). To assess the differences between the percentages of the GroEL-GroES complexes containing two GFP molecules, statistical analysis was performed using Student's t test. The asterisk indicates p < 0.01.