Background: The mismatch recognition complex Msh2-Msh6 undergoes an ATP-mediated conformational change.
Results: An engineered disulfide bond inhibits the conformational change and ATP-induced formation of the mispair-dependent Msh2-Msh6 sliding clamp and Msh2-Msh6-Mlh-Pms1 ternary complexes.
Conclusion: The conformational change is required for mismatch repair.
Significance: This is the first direct test for the requirement of the Msh2-Msh6 conformational change in mismatch repair.
Keywords: DNA Mismatch Repair, DNA Repair, Enzyme Mechanisms, Molecular Genetics, Molecular Modeling
Abstract
ATP binding causes the mispair-bound Msh2-Msh6 mismatch recognition complex to slide along the DNA away from the mismatch, and ATP is required for the mispair-dependent interaction between Msh2-Msh6 and Mlh1-Pms1. It has been inferred from these observations that ATP induces conformational changes in Msh2-Msh6; however, the nature of these conformational changes and their requirement in mismatch repair are poorly understood. Here we show that ATP induces a conformational change within the C-terminal region of Msh6 that protects the trypsin cleavage site after Msh6 residue Arg1124. An engineered disulfide bond within this region prevented the ATP-driven conformational change and resulted in an Msh2-Msh6 complex that bound mispaired bases but could not form sliding clamps or bind Mlh1-Pms1. The engineered disulfide bond also reduced mismatch repair efficiency in vivo, indicating that this ATP-driven conformational change plays a role in mismatch repair.
Introduction
Mismatch repair (MMR)2 identifies and repairs mispaired bases that occur in DNA as the result of errors during DNA replication, the formation of recombination intermediates, or chemical damage to DNA (1–5). As MMR normally functions to suppress the accumulation of mutations, defects in MMR genes underlie both inherited and spontaneous cancers that are driven by high rates of accumulating mutations (6–9). In Escherichia coli, the MutS homodimer recognizes mispairs (2, 3, 10–13), whereas in Saccharomyces cerevisiae and humans, one of two heterodimers of MutS homologs, Msh2-Msh3 or Msh2-Msh6, recognizes mispairs (4, 14–17). Mispair recognition by the MutS family of proteins is the first step of MMR that leads to the recruitment of either the bacterial MutL homodimer or the eukaryotic MutL homolog heterodimers, Mlh1-Pms1 or Mlh1-Mlh3, to mediate downstream steps, which lead to excision and resynthesis of the DNA strand that contained the misincorporated bases (18–26).
MutS and its homologs contain ABC ATPase domains, and multiple lines of evidence indicate that ATP plays crucial roles in the function of these proteins (27–36). In the absence of nucleotide or the presence of ADP, these complexes can recognize mispairs; however, these pre-bound complexes form “sliding clamps” that dissociate from the mispair and slide along the DNA when challenged with ATP or nonhydrolyzable ATP analogs (18, 19, 28, 30). These sliding clamps can be trapped on DNA fragments with sterically blocked ends (18, 19, 24, 30, 37). ATP binding by MutS and its homologs is also required for the ability of these proteins to interact with MutL or the eukaryotic MutL homologs, respectively, and thereby mediate downstream events in MMR (18–20, 24). Together these biochemical results are consistent with the presence of an ATP-induced conformational change in MutS, Msh2-Msh3, and Msh2-Msh6 that propagates from the ATPase domain to other parts of the molecule.
To date, the details of ATP-driven conformational changes in MutS, Msh2-Msh3, and Msh2-Msh6 have proven elusive (38). Crystal structures of bacterial MutS, human Msh2-Msh3, and Msh2-Msh6 bound to mispairs have been solved and have provided important details of the ADP- and mispair-bound states of these proteins (2, 10, 39, 40). This state has been called the open form (41, 42). In contrast, the conformation of the ATP-bound forms of these proteins is unknown. Based on structural homology to ATP-bound Rad50 (43), this form of the MutS family of proteins has been called the closed form (41, 42). Consistent with these inferred states, deuterium exchange studies with S. cerevisiae Msh2-Msh6 demonstrated that ATP binding protects the Msh2-Msh6 dimer interface between the ATPase domains and exposes the mispair-binding domains (42). ATP binding moderately increases deuterium exchange in the connector domains of MutS and Msh2 (42), which interact with MutL or Mlh1-Pms1, respectively (44, 45). These data are consistent with a model in which ATP binding releases the mispair-binding domain from the mispair and exposes the connector domain to recruit MutL/Mlh1-Pms1. However, the details of and the requirement for these conformational changes in MMR remain poorly characterized. In the studies presented here, we have used a combination of genetic and biochemical approaches to identify a region undergoing an ATP-induced conformational change and demonstrate its functional involvement in MMR.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Genetic Manipulations
S. cerevisiae strains were grown in standard medium, including either yeast extract/peptone/dextrose or complete supplement mixture medium (U.S. Biological Corp.) lacking specific amino acids to select for plasmid markers and/or Thr+ revertants. E. coli strains used for the propagation of plasmids were grown in LB medium containing 100 μg/ml of ampicillin to select for plasmids. All S. cerevisiae strains used for genetic tests were isogenic derivatives of the same parental S288C strain (46). The strain used for the complementation studies was RDKY4234 (MATα ura3–52 leu2Δ1 trp1Δ63 his3Δ200 hom3–10 lys2::InsE-A10 msh3Δ::hisG msh6Δ::hisG). Plasmid pRDK439 was from our laboratory collection and was constructed by cloning a 6-kb fragment of S. cerevisiae genomic DNA containing MSH6 into the ARS CEN vector pRS315. The dual Msh2-Msh6 E. coli expression plasmid pET11a-Msh2-Msh6 was from Dr. Manju Hingorani (47). Derivatives of pRDK439 and pET11a-Msh2-Msh6 containing the indicated MSH6 alleles were made by site-directed mutagenesis and verified by sequencing the complete MSH6 gene (Table 1). The msh6 truncation alleles 1125stop, 1135stop, 1152stop, 1172stop, 1182stop, 1191stop, 1201stop, 1211stop, 1221stop, and 1241stop were engineered by inserting two stop codons at the indicated positions. The msh6 truncation alleles 1231stop and 1236stop were engineered by deleting codons 1231–1242 and 1236–1242, respectively. All of the genetic methods used in the described studies, including those used for site-directed mutagenesis, verification by DNA sequencing, and evaluation of mutator phenotypes by patch tests and fluctuation analysis, were as described previously (15, 35, 48, 49).
TABLE 1.
Plasmids
| Plasmid name | Description |
|---|---|
| pRDK439 | pRS315-MSH6 |
| pRDK1641 | pRS315-msh6 1125stop |
| pRDK1642 | pRS315-msh6 1135stop |
| pRDK1643 | pRS315-msh6 1152stop |
| pRDK1644 | pRS315-msh6 1172stop |
| pRDK1645 | pRS315-msh6 1182stop |
| pRDK1646 | pRS315-msh6 1191stop |
| pRDK1647 | pRS315-msh6 1201stop |
| pRDK1648 | pRS315-msh6 1211stop |
| pRDK1649 | pRS315-msh6 1221stop |
| pRDK1650 | pRS315-msh6 1231stop |
| pRDK1651 | pRS315-msh6 1236stop |
| pRDK1652 | pRS315-msh6 1241stop |
| pRDK1653 | pRS315-msh6-L1129D |
| pRDK1654 | pRS315-msh6-S1137A |
| pRDK1655 | pRS315-msh6-L1129D S1137A |
| pRDK1656 | pRS315-msh6-L1129C |
| pRDK1657 | pRS315-msh6-S1137C |
| pRDK1658 | pRS315-msh6-L1129C S1137C |
| pET11a-MSH2-MSH6 | pET11a-MSH2-MSH6 |
| pRDK1640 | pET11a-MSH2-msh6-L1129C S1137C |
| pRDK573 | pRS424-GAL-MLH1 |
| pRDK1099 | pRS425-GAL-PMS1-FLAG |
Proteins
Wild-type and mutant Msh2-Msh6 complexes were purified from E. coli as described (24, 47, 49, 50). Wild-type Msh2-Msh6 and Msh2-Msh6 L1129C/S1137C were purified in buffer A (25 mm Tris, pH 8.0, 314 mm NaCl, 1 mm EDTA, 10% glycerol, 5 mm DTT, 0.02% Igepal, 1 mm phenylmethylsulfonyl fluoride (PMSF), 1 mm benzamidine, 0.5 mg/liter of bestatin, and 1 mg/liter each of chymostatin, pepstatin A, aprotinin, and leupeptin). Reduced wild-type Msh2-Msh6 and reduced Msh2-Msh6 L1129C/S1137C were maintained in buffer A. Wild-type Msh2-Msh6 and Msh2-Msh6 L1129C/S1137C were oxidized by overnight dialysis at 4 °C against buffer A lacking DTT. ATP-supplemented oxidized Msh2-Msh6 L1129C/S1137C was generated by overnight dialysis at 4 °C against buffer A lacking DTT and containing 100 μm ATP in the absence of Mg2+. Mlh1-Pms1 was purified from S. cerevisiae as described (24, 41). LacI protein was generously provided by Kathleen Matthews (Rice University).
Mass Spectrometry
To determine which region of Msh6 was undergoing the ATPγS-induced conformational change, ∼300 ng each of full-length Msh6 and the 90-, 80-, and 75-kDa Msh6 tryptic fragments were separated by SDS-PAGE on a 4–15% gradient gel. After staining and destaining, the gel was incubated in water overnight, and the bands of interest were each excised from the gel with a clean razor blade. In gel slice reduction, alkylation, and trypsin digestion was performed by the Proteomics Mass Spectrometry Laboratory at the Scripps Research Institute, La Jolla, CA. Briefly, the proteins were reduced in 10 mm DTT (Sigma) for 1 h, then alkylated with 55 mm iodoacetamide (Sigma) for 30 min in the dark, and finally digested for 18 h with trypsin at 37 °C using a 1:30 (w/w) enzyme to substrate ratio. The digestion reaction was stopped by lowering the pH of the reaction to 2.0. Tryptic peptides were analyzed by reverse phase chromatography using Zorbax SB-C18 stationary phase (Agilent). The data-dependent MS/MS data were obtained on a LTQ linear ion trap mass spectrometer (Thermo Scientific). Protein identification was performed using Mascot (Matrix Science, version 2.1.04). Tandem mass spectra were extracted by the Xcalibur software, and all MS/MS samples were analyzed using Mascot. Scaffold (Proteome Software Inc.) was used to additionally validate MS/MS-based peptide and protein identifications.
To demonstrate the presence of the disulfide bond, 20 μg of oxidized and reduced Msh2-Msh6 L1129C/S1137C were each treated with 2% iodoethanol (Sigma) for 1 h at 37 °C in the dark. The protein was then precipitated by addition of a mixture of 50% ethanol, 50% acetone containing 0.1% acetic acid for 1 h on ice. Then the proteins were denatured with 8 m urea, reduced with 10 mm DTT for 10 min at 42 °C, alkylated with 30 mm iodoacetamide (Sigma) for 30 min at room temperature in the dark, digested with 0.2 μg of sequencing grade trypsin (Promega) at 37 °C overnight, and the digestion reaction was stopped with 0.2% trifluoroacetic acid. Peptide cleanup was then performed using a 50-mg Sep-Pak C18 column (Waters) that had been equilibrated with 0.1% acetic acid. Bound peptides were eluted from the column with 300 μl of 80% acetonitrile, 0.1% acetic acid; the eluate was lyophilized; and the resulting peptides were resuspended in 0.1% trifluoroacetic acid. The samples were then analyzed by LC-MS/MS using an LTQ Orbitrap Discovery XL mass spectrometer (Thermo Scientific). Data dependent MS/MS data collected were searched using SorcererTM-SEQUEST (51) with a semi-tryptic modified database designed for the mutant Msh6. Variable modifications for cysteine were applied for carbamidomethyl (+57 Da) and 2-hydroxyethyl (+44 Da) with a peptide mass tolerance of 20 ppm. MS/MS peptide modification and protein identification were additionally validated by manual inspection.
Partial Proteolysis
To monitor the ATPγS-induced conformational change in Msh2-Msh6, 4 μg of wild-type Msh2-Msh6 or Msh2-Msh6 L1129C/S1137C was incubated with 0–500 ng of trypsin with or without 100 μm ATPγS in reaction buffer (25 mm Tris, pH 8.0, 110 mm NaCl, 4 mm MgCl2, 0.01% Igepal, 2 mm dithiothreitol, 2% glycerol) in a final volume of 10 μl for 1 h at room temperature (23 °C). Reactions were stopped by the addition of PMSF to a final concentration of 10 mm. The proteolysis products were separated by SDS-PAGE on a 4–15% gel (Bio-Rad), and the resulting gel was silver-stained.
Surface Plasmon Resonance
Experiments analyzing the interaction between the Msh2-Msh6 complex and mispaired DNA as well as the formation of the Msh2-Msh6-Mlh1-Pms1 ternary complex on mispaired DNA were performed with a Biacore T100 instrument (GE Healthcare) essentially as described previously (41, 44). Note that the buffers used in these experiments contained Mg2+.
UV Cross-linking
Nucleotide binding was measured by UV cross-linking essentially as described previously (28). Briefly, 20-μl reactions containing 10 pmol of reduced wild-type Msh2-Msh6, 30 pmol of oxidized wild-type Msh2-Msh6, 30 pmol of reduced Msh2-Msh6 L1129C/S1137C, 50 pmol of oxidized Msh2-Msh6 L1129C/S1137C, or 50 pmol of oxidized Msh2-Msh6 L1129C/S1137C that had been dialyzed overnight against buffer containing ATP without Mg2+, were each performed in binding buffer that contained 50 mm Tris, pH 8.0, 110 mm NaCl, 100 μg/ml of BSA, 0.5 mm EDTA, 5% glycerol. The reactions with reduced protein also contained 2 mm dithiothreitol. The absence of magnesium prevented ATP hydrolysis. Proteins were mixed with 25 Ci/mmol of [γ-32P]ATP and incubated on ice for 10 min. Samples were then subjected to UV cross-linking (Stratalinker) for 20 min followed immediately by fractionation by SDS-PAGE using 4–15% gradient gels. The gels were exposed to a PhosphorImager screen overnight, and the screen was developed using a PhosphorImager (GE Healthcare).
RESULTS
ATP Binding to Msh2-Msh6 Protects the C Terminus of Msh6 from Trypsin Cleavage
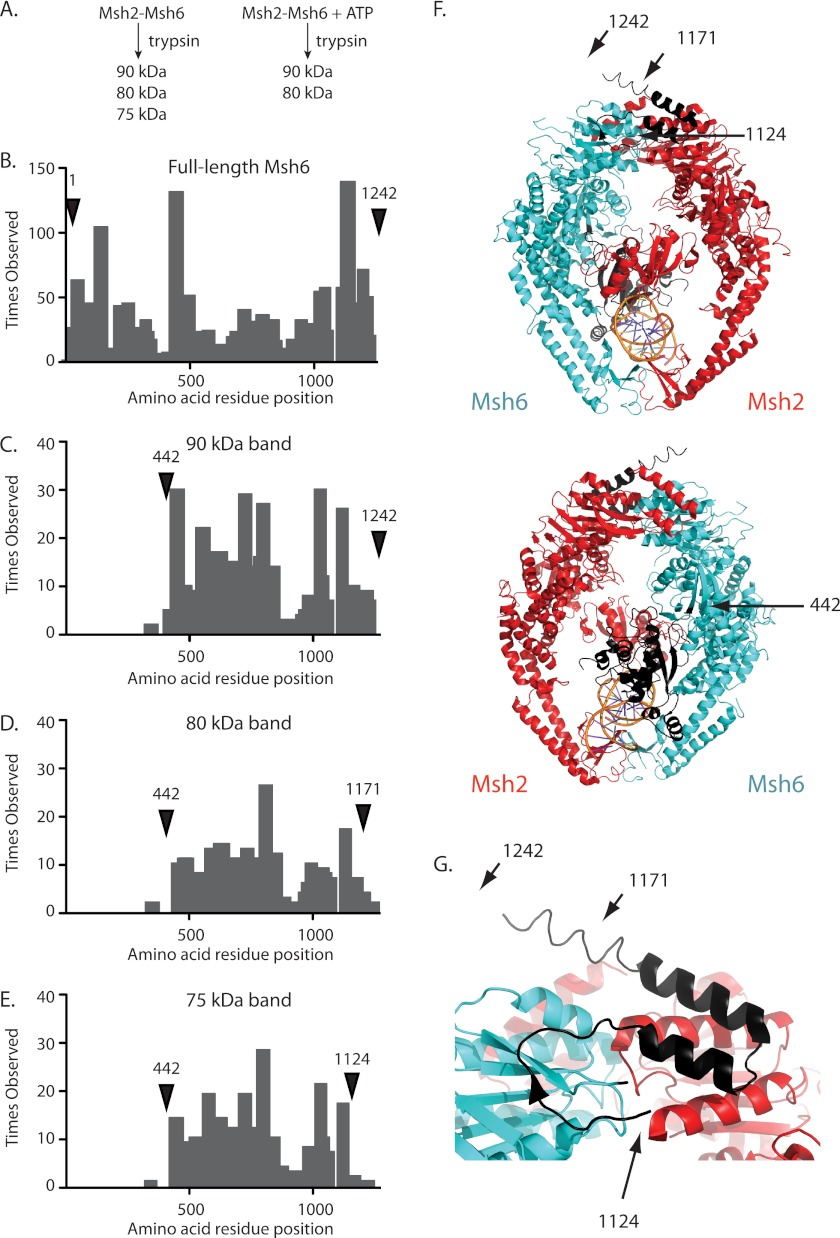
We previously demonstrated that ATP binding protected Msh2-Msh6 from digestion with trypsin (Fig. 1A), consistent with a conformational change (41). To determine which part of Msh2-Msh6 undergoes the ATP-induced conformational change, bands corresponding to full-length Msh6 and the 90-, 80-, and 75-kDa trypsin fragments were excised from a gel, digested to completion with trypsin, and analyzed by mass spectrometry (MS; Fig. 1, B–E). Consistent with Western blots (41), MS confirmed that the 90-, 80-, and 75-kDa trypsin fragments were derived from Msh6.
FIGURE 1.
Identification of the ATP-protected trypsin site in Msh6. A, the addition of ATP to a partial proteolysis reaction with Msh2-Msh6 prevents the generation of the 75-kDa fragment as previously demonstrated (41). B–E, histograms of the number of times each amino acid was observed by mass spectrometry for (B) full-length Msh6 and the (C) 90, (D) 80, and (E) 75-kDa trypsin fragments. Labeled arrows indicate the predicted N and C termini for each trypsin fragment. F, ribbon diagrams of human Msh2-Msh6 and bound mispaired DNA (Msh2, red; Msh6, blue; DNA, orange; PDB code 2O8B (39)) and the same structure rotated 180° generated with PyMol (59). The positions of the N and C termini of the 90-kDa fragment (residues 442–1242), 80-kDa fragment (residues 442–1171), and 75-kDa fragment (residues 442–1124) are indicated with arrows to the equivalent residues in human Msh6. G, expanded view of Msh6 C terminus with the C termini of the 90-, 80-, and 75-kDa fragments indicated with arrows.
The 90-, 80-, and 75-kDa fragments yielded the same pattern and proportions of N-terminal tryptic peptides, and therefore appeared to have the same N terminus. For each trypsin fragment, MS failed to detect significant amounts of any peptides N-terminal to the trypsin cleavage site at Arg441-Glu442 (Fig. 1, C–E), unlike full-length Msh6 from which substantial amounts of peptides starting with the first residue were detected (Fig. 1B). This suggests that the N terminus of the 90-, 80-, and 75-kDa fragments was residue Glu442. Residue Glu442 lies in the N terminus of the connector domain of Msh6, C-terminal to a loop connecting the connector domain with the mispair-binding domain and appears to be exposed. Thus, the 90-, 80-, and 75-kDa fragments lack the mispair-binding domain and the N-terminal tether that contains the PCNA interaction site (Fig. 1, C–E).
MS analysis indicated the 90-, 80-, and 75-kDa fragments had different C termini, which were consistent with their molecular weights estimated by SDS-PAGE. As for intact Msh6, significant amounts of peptides from the 90-kDa fragment sample were observed up to and including the C-terminal peptide ending in residue Ser1242 (Fig. 1C). An Msh6 Glu442-Ser1242 fragment has a calculated molecular mass of 90.4 kDa, consistent with the estimated size of the fragment analyzed. In contrast, the 80-kDa fragment had reduced levels of tryptic peptides between residues Arg1171 and Ser1242 (Fig. 1D). Residue Arg1171 is in the last α-helix of the ATPase domain of Msh6, as predicted by homology to the human Msh2-Msh6 structure (Fig. 1, F and G), and an Msh6 Glu442-Arg1171 fragment has a calculated molecular mass of 82.7 kDa, consistent with the estimated size of the fragment analyzed. MS analysis of the 75-kDa fragment was indicative of an even smaller fragment; significant levels of tryptic peptides were not identified C-terminal to residue Arg1124 (Fig. 1E). Residue Arg1124 is between the last two β strands of the ATPase domain, as predicted by homology to the human Msh2-Msh6 structure (Fig. 1, F and G), and an Msh6 Glu442-Arg1124 fragment has a calculated molecular mass of 77.5 kDa, consistent with the estimated size of the fragment analyzed. These results support the idea that ATP binding induces a conformational change in the region containing Msh6 residue Arg1124, which protects the Arg1124-Asn1125 trypsin site from cleavage.
The C Terminus of Msh6 Is Required for MMR
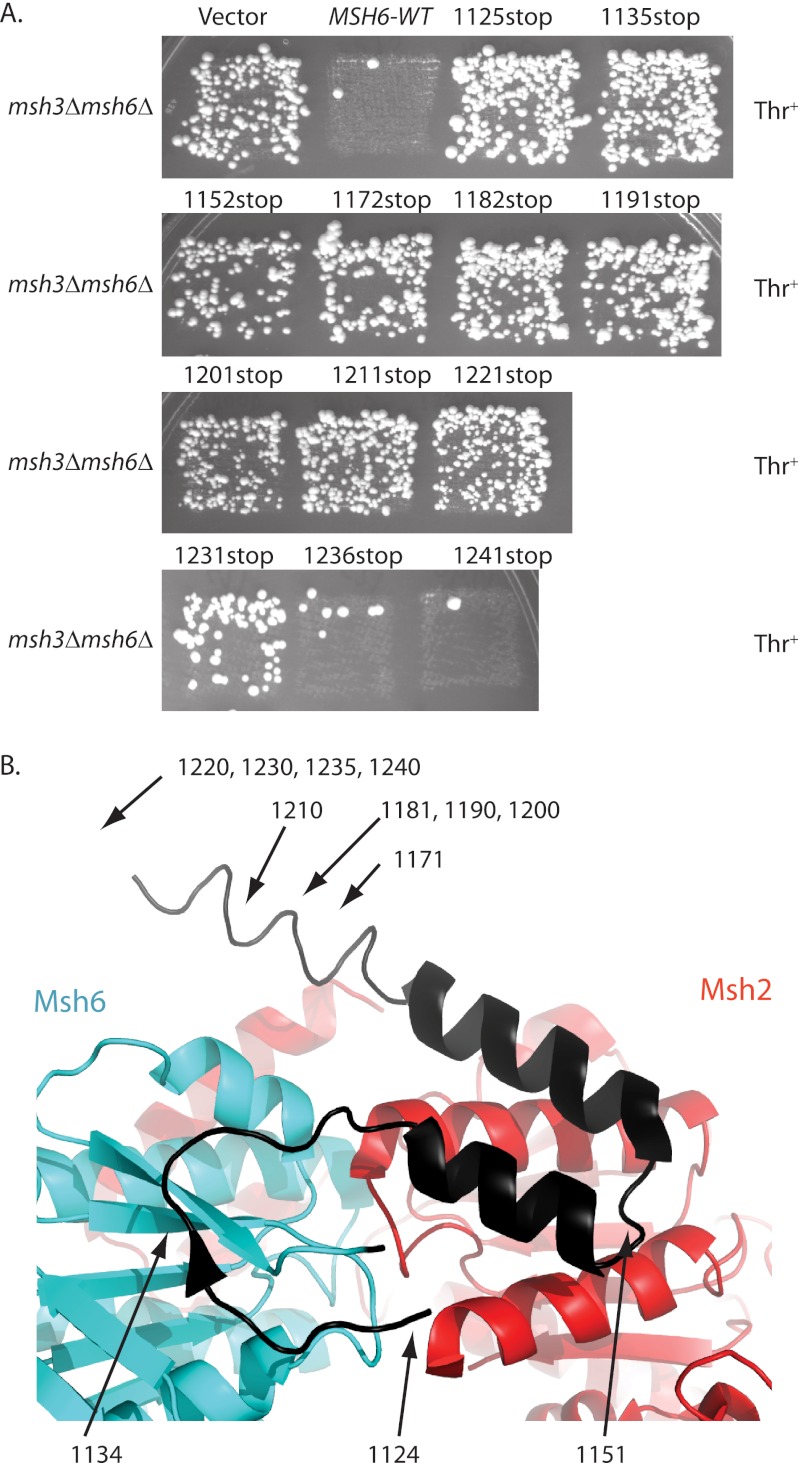
To determine whether the C-terminal region of Msh6 that is affected by ATP binding is required for MMR, we constructed a series of MSH6 mutations that eliminated amino acids from the C terminus of Msh6. These mutant genes were expressed on low copy number plasmids and tested for their ability to complement the high frameshift mutation rate of an msh3Δ msh6Δ strain. The mutant lacking the last two codons of MSH6 (1241stop) fully complemented the high frameshift mutation rate of an msh3Δ msh6Δ strain in patch tests, like a wild-type MSH6 plasmid (Fig. 2A), and had a similar mutation rate to that of the wild-type MSH6 plasmid in quantitative mutation rate tests (Table 2). The mutant lacking the last seven codons of MSH6 (1236stop) had an intermediate mutation rate, which was 8-fold higher than seen with the wild-type MSH6 plasmid (Table 2). In contrast, deletion of 12 or more amino acids from the C terminus (1125stop to 1231stop) resulted in plasmids that failed to complement the high frameshift mutation rate of an msh3Δ msh6Δ strain in patch tests, like the empty vector control plasmid (Fig. 2A). The strains containing plasmids with stop codons at 1221 and 1231 had mutation rates that were 199- and 149-fold higher, respectively, than that obtained with a plasmid containing wild-type MSH6, and were similar to the rates obtained with the empty vector control plasmid (Table 2). These results indicate that virtually the entire C-terminal region of Msh6 is required for MMR, including regions not ordered in the human Msh2-Msh6 structure (Fig. 2B) (39), and would be consistent with a C-terminal Msh2-Msh6 dimerization domain equivalent to the human Msh2-Msh3 C-terminal domain (40).
FIGURE 2.
Msh6 residues 1124–1230 are required for MMR. A, patches of msh3Δ msh6Δ strains expressing plasmid-borne msh6 truncation alleles (Table 1) were replica plated onto plates lacking leucine and threonine to test for MMR complementation by suppression of hom3–10 reversion. The positions of the engineered stop codons are indicated. B, a ribbon diagram of human Msh2-Msh6 with the C terminus of Msh6 in black. The positions of the last codons in S. cerevisiae Msh6 are indicated with arrows (S. cerevisiae Msh6 residues 1171–1207 are missing from human Msh6 and the equivalent to S. cerevisiae Msh6 residue 1214 is the last ordered residue in the human Msh2-Msh6 structure).
TABLE 2.
Mutation rate analysis of msh6 mutants
| Relevant Genotype | hom3–10 reversion rate (fold-increase) |
|---|---|
| msh3Δ MSH6 | 3.72 (1.95–13.4) × 10−8 (1) |
| msh3Δ msh6Δ | 4.96 (3.53–9.99) × 10−6 (133) |
| msh3Δ msh6 1221 stop | 7.39 (3.58–13.8) × 10−6 (199) |
| msh3Δ msh6 1231 stop | 5.55 (2.06–11.5) × 10−6 (149) |
| msh3Δ msh6 1236 stop | 2.96 (0.96–7.41) × 10−7 (8.0) |
| msh3Δ msh6 1241 stop | 2.60 (1.82–5.45) × 10−8 (0.7) |
| msh3Δ msh6-L1129C | 9.73 (6.96–28.1) × 10−8 (2.6) |
| msh3Δ msh6-S1137C | 4.67 (3.6–16.8) × 10−8 (1.3) |
| msh3Δ msh6-L1129C S1137C | 3.16 (2.36–10.6) × 10−7 (8.5) |
An Engineered Disulfide Bond in the C Terminus of Msh6 Disrupts MMR
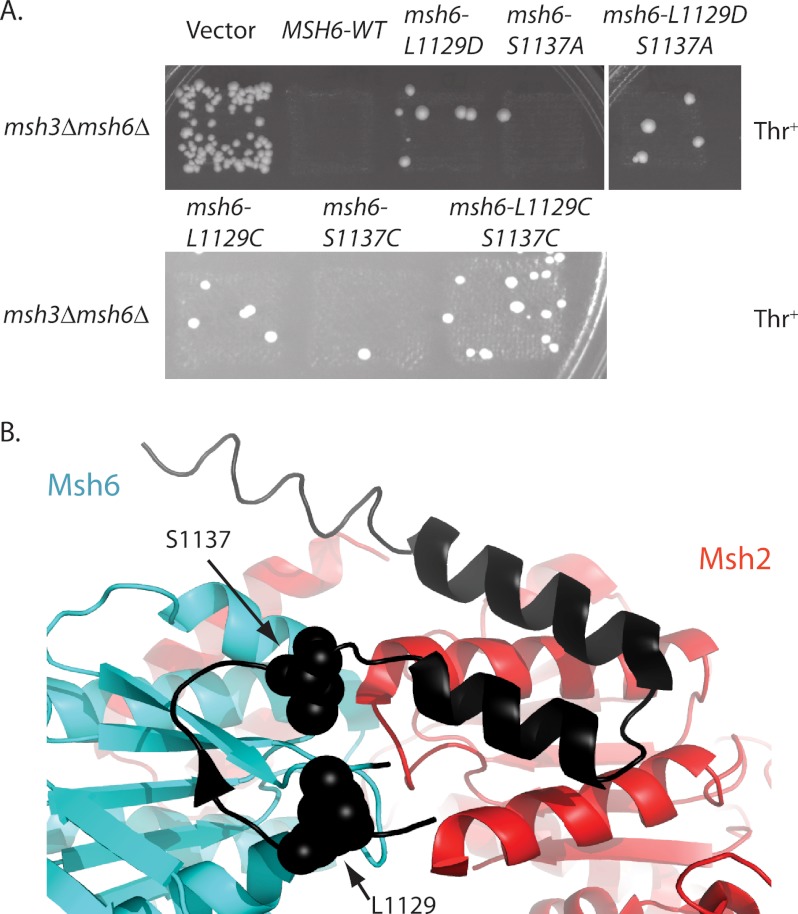
We next analyzed the C-terminal region of Msh6 near the ATP-protected Arg1124-Asn1125 trypsin site for residues that could be changed to cysteines that would then have the potential to generate a disulfide bond under oxidizing conditions. Residues 1129 and 1137 are in close proximity and are in a region undergoing ATP-induced changes (42), but they are not ideally positioned for disulfide bond formation in the crystal structure of human Msh2-Msh6 (39). We reasoned that if conformational changes allowed for disulfide bond formation in the mutant protein, then disulfide bond formation might in turn affect conformational changes in Msh2-Msh6.
We then created single and double cysteine mutations in a plasmid-encoded MSH6 gene and tested for their effect on MMR (Fig. 3A). The msh6-L1129C and msh6-S1137C mutations individually appeared to be weak mutator mutations in patch tests as did alternative mutations affecting these residues including msh6-L1129D and msh6-S1137A. In contrast, the msh6-L1129C/S1137C pair of mutations, which we predicted to form a disulfide bond, caused a significantly stronger mutator phenotype in patch tests, unlike the msh6-L1129D/S1137A pair. Other potential cysteine pairs in this region were investigated but not studied further as the other double cysteine mutants did not have increased mutation rates over the single cysteine mutants. Quantitative mutation rate tests confirmed that the msh6-L1129C/S1137C allele caused a significant increase in mutation rate although not as large as a complete deletion of MSH6 (Table 2). In contrast, the mutation rates caused by the msh6-L1129C and msh6-S1137C mutations were not significantly different from the wild-type mutation rate (Table 2). Thus, the MMR defect occurred only in the double cysteine mutant.
FIGURE 3.
msh2-msh6 L1129C/S1137C causes an elevated mutation rate. A, patches of msh3Δ msh6Δ strains expressing plasmid-borne msh6 alleles (Table 1) were replica plated onto plates lacking leucine and threonine to test for MMR complementation by suppression of hom3–10 reversion. B, residues equivalent to S. cerevisiae Msh6 Leu1129 and Ser1137 are depicted by spheres on the ribbon diagram of human Msh2-Msh6 with the C terminus of Msh6 in black.
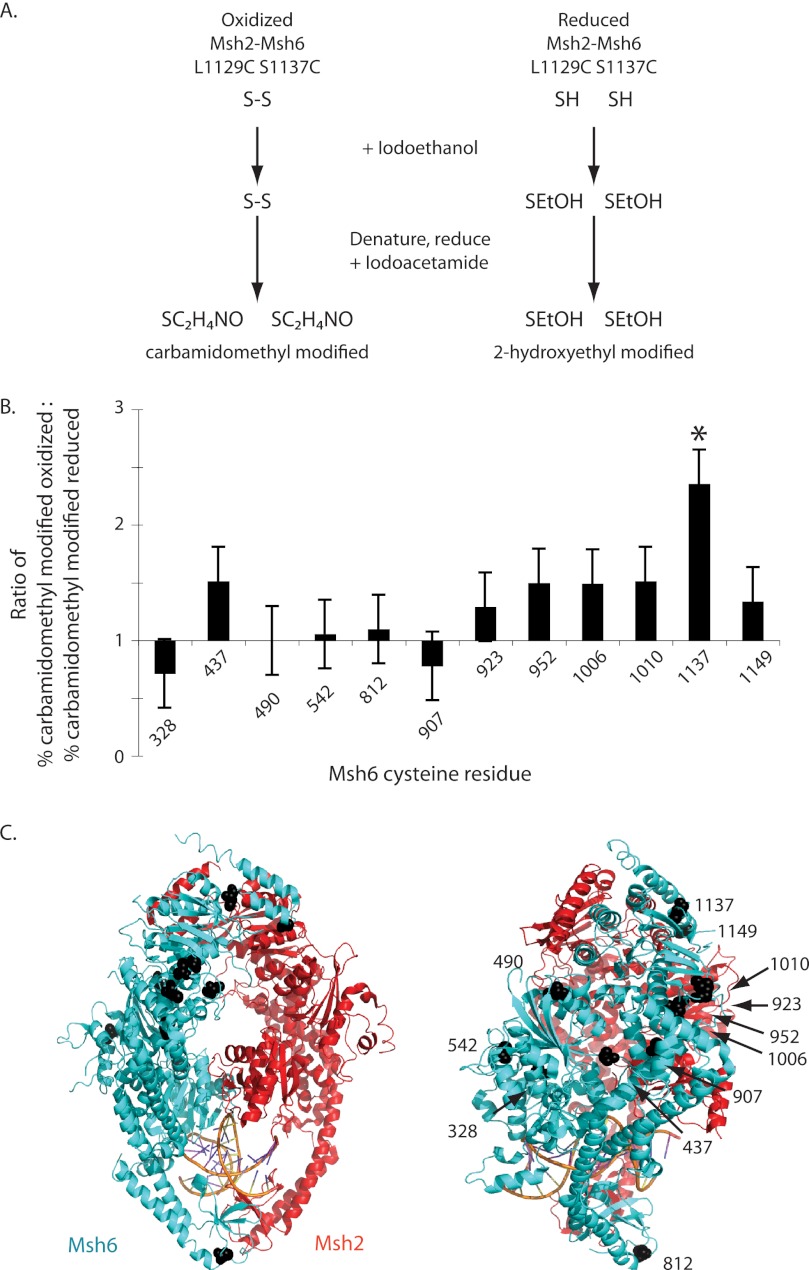
To demonstrate the formation of a disulfide bond, Msh2-Msh6 L1129C/S1137C was overexpressed and purified. An oxidized sample was generated by overnight dialysis against buffer A lacking DTT to enable any potential disulfide bonds to form. Both oxidized and reduced Msh2-Msh6 L1129C/S1137C were treated with iodoethanol, denatured, reduced, and treated with iodoacetamide. In this experiment, exposed cysteines not in disulfide bonds will react with iodoethanol to become 2-hydroxyethyl-modified and have a total mass of 147 Da. Cysteines in disulfide bonds or buried within the protein will react with iodoacetamide after denaturation and reduction to become carbamidomethyl-modified and have a total mass of 160 Da (Fig. 4A). Thus, exposed cysteines in disulfide bonds should be 2-hydroxyethyl-modified in the reduced sample and carbamidomethyl-modified in the oxidized sample. In contrast, cysteines not in disufide bonds should be either 2-hydroxyethyl-modified if they are exposed or carbamidomethyl-modified if they are buried regardless of whether the sample is oxidized or reduced.
FIGURE 4.
Residues L1129C and S1137C form a disulfide bond in oxidized Msh2-Msh6 L1129C/S1137C. A, schematic of the alkylation reactions. B, ratio of carbamidomethylation in the oxidized sample to carbamidomethylation in the reduced sample for Msh6 L1129C/S1137C cysteines. The error bars indicate the S.D. C, residues equivalent to the cysteines in S. cerevisiae Msh6 are depicted by black spheres on the ribbon diagram human Msh2-Msh6.
After this treatment, S1137C was observed as carbamidomethyl-modified 73% of the time in the oxidized sample and 31% in the reduced sample (Fig. 4B). L1129C was not observed in the oxidized sample and was only observed once in the reduced sample. L1129C may not have been detected for a variety of technical reasons including vaporization and/or time of flight. Thus no significant conclusions can be drawn about L1129C. However, it should be noted that this analysis method allows inference of disulfide bond status even when both cysteine-containing peptides are not recovered as disulfide formation is detected through analysis of the properties of the modified cysteines. Eleven additional Msh6 cysteines were observed more than once in both the oxidized and reduced samples (Fig. 4C), but only the modification of S1137C was significantly affected by the presence of DTT (p = 0.003; Fisher's exact test). Thus, at least 73% of S1137C in oxidized Msh2-Msh6 L1129C/S1137C was disulfide bonded. Possible explanations for the observation of 27% 2-hydroxyethyl-modified S1137C are incomplete disulfide bond formation due to protein dynamics in this region of the protein or that dialysis treatment was insufficient to oxidize 100% of the cysteine residues.
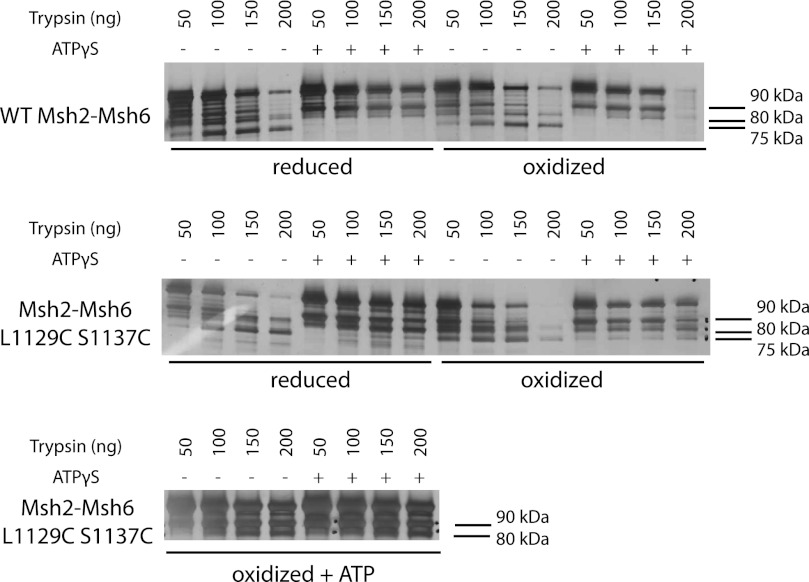
Oxidized Msh6 L1129C/S1137C Prevents ATP-induced Conformational Changes
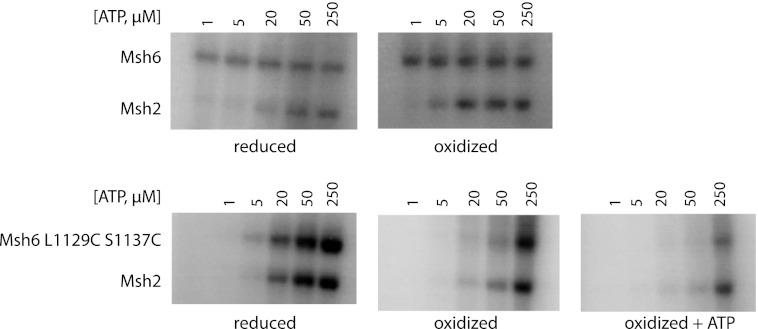
To determine how the engineered disulfide bond affected the ATP-induced Msh2-Msh6 conformational change, we repeated the partial proteolysis analysis using reduced and oxidized Msh2-Msh6 L1129C/S1137C. As for both reduced and oxidized wild-type Msh2-Msh6, ATPγS prevented trypsin from cleaving the reduced Msh6 L1129C/S1137C into the 75-kDa fragment (Fig. 5). In contrast, trypsin generated the 75-kDa fragment from oxidized Msh2-Msh6 L1129C/S1137C regardless of whether or not ATPγS was present, consistent with the possibility that disulfide bond formation trapped Msh2-Msh6 L1129C/S1137C in the ATP-unbound conformation. When the oxidized Msh2-Msh6 L1129C/S1137C was dialyzed overnight against buffer lacking both DTT and Mg2+ and containing 100 μm ATP at 4 °C, trypsin digestion was unable to generate the 75-kDa fragment, regardless of whether or not ATPγS was in the proteolysis reaction (Fig. 5), consistent with slow binding of ATP and subsequent conformational changes during dialysis. Together, these results suggest that oxidized Msh2-Msh6 L1129C/S1137C can bind ATP and undergo ATP-driven conformational changes but on an extremely slow time scale.
FIGURE 5.
Oxidized Msh2-Msh6 L1129C/S1137C is trapped in the nucleotide-free conformation. Four μg of reduced or oxidized wild-type Msh2-Msh6 or reduced, oxidized, or ATP-supplemented oxidized Msh2-Msh6 L1129C/S1137C were incubated with 50–200 ng of trypsin with or without 100 μm ATPγS. The proteolysis products were separated by SDS-PAGE on a 4–15% gel, and the gel was silver stained. Only the portion of the gels containing the 70–100-kDa size range is shown.
Oxidized Msh2-Msh6 L1129C/S1137C Binds Mispaired Bases But Is Defective for Sliding Clamp Formation
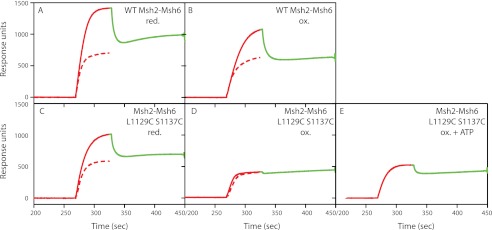
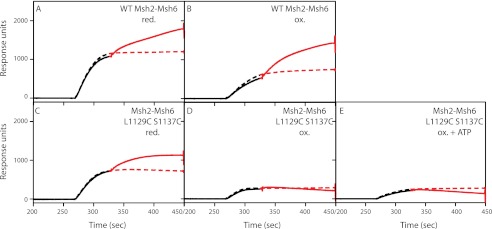
Wild-type Msh2-Msh6 in either the nucleotide free or ADP-bound state can bind to mispairs. Challenging these complexes with ATP converts wild-type Msh2-Msh6 to a sliding clamp that slides away from the mispair along the DNA (28). Sliding clamp formation by Msh2-Msh6 can be monitored by surface plasmon resonance by following the accumulation of multiple Msh2-Msh6 molecules on an immobilized DNA molecule containing a single mispair under conditions in which the DNA ends are blocked (24). To do this, a 236-base pair oligonucleotide with a central GT mispair, a biotin at one end, and a lac operator sequence at the other end was bound to a streptavidin-coated Biacore chip at its biotinylated end. Addition of LacI, which binds the lac operator and can be removed by addition of isopropyl 1-thio-β-d-galactopyranoside, sterically blocks the DNA end and prevents Msh2-Msh6 from sliding off of the DNA.
Both oxidized and reduced wild-type Msh2-Msh6 bound mispaired DNA in the presence of ATP and Mg2+ (Fig. 6, A and C, red dashed line). Under these conditions, free Msh2-Msh6 hydrolyzes ATP to yield the ADP-bound state in which it binds the mispair, exchanges ADP for ATP, and then slides off the DNA end, resulting in the steady-state level of binding seen (24). Addition of LacI increases the number of molecules bound to the DNA substrate (Fig. 6, red solid line), and release of LacI with isopropyl 1-thio-β-d-galactopyranoside returns binding back to the steady-state level (Fig. 6, green dashed line). This pattern of LacI-dependent accumulation of Msh2-Msh6 on the DNA substrate demonstrated that both oxidized and reduced wild-type Msh2-Msh6 can form sliding clamps (24). Reduced Msh2-Msh6 L1129C/S1137C had the same binding properties as wild-type Msh2-Msh6, indicating that the mutant can form sliding clamps under reduced conditions (Fig. 6B). In contrast, oxidized Msh2-Msh6 L1129C/S1137C (Fig. 6D) and oxidized Msh2-Msh6 L1129C/S1137C that had been dialyzed overnight against buffer containing ATP (Fig. 6E) were able to bind mispaired bases (Fig. 7), but appeared to be defective for the formation of sliding clamps as they showed no increase in binding to substrates with blocked ends versus unblocked ends, and there was little to no decrease in binding to the end-blocked substrate upon addition of isopropyl 1-thio-β-d-galactopyranoside. It should be noted that ATP-supplemented oxidized Msh2-Msh6 L1129C/S1137C can bind to the mispaired base because the Biacore running buffer contains Mg2+, which allows the bound ATP to be hydrolyzed, creating a protein state that is permissible for mispair binding (24, 28). Therefore, the inability of oxidized Msh2-Msh6 L1129C/S1137C and ATP-supplemented oxidized Msh2-Msh6 L1129C/S1137C to form a sliding clamp suggests that disulfide bond formation completely or partially inhibits ATP-induced conformational changes.
FIGURE 6.
Msh2-Msh6 L1129C/S1137C cannot form a sliding clamp under oxidizing conditions but can form a sliding clamp under reducing conditions. Approximately 20 ng of mispair-containing DNA was conjugated to a flow cell of a streptavidin-coated Biacore SA chip. In the unblocked sliding experiments, depicted by the red dashed line, 50 nm wild-type Msh2-Msh6 or Msh2-Msh6 L1129C/S1137C and 250 μm ATP were flowed over the chip. In the end-blocked sliding experiments, depicted by the red solid line, 30 nm LacI was flowed over the chip in reaction buffer before and during Msh2-Msh6 binding. Dissociation of Msh2-Msh6 from the DNA end was induced by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside to the sample, which is depicted by the green solid line.
FIGURE 7.
Oxidized Msh2-Msh6 L1129C/S1137C prefers binding mispaired DNA to base-paired DNA. Binding of Msh2-Msh6 complexes to a 236-bp DNA containing a central GT mispair or GC base pair in the presence of 250 μm ATP was analyzed on a BIAcore T100 biosensor. The solid black line indicates association of the indicated Msh2-Msh6 complex with a fully base-paired DNA substrate. The solid red line indicates association with a DNA substrate containing a central GT mispair. All DNA substrates had LacI end blocks. A, reduced wild-type Msh2-Msh6; B, reduced Msh2-Msh6 L1129C/S1137C; C, oxidized wild-type Msh2-Msh6; D, oxidized Msh2-Msh6 L1129C/S1137C; E, ATP-supplemented oxidized Msh2-Msh6 L1129C/S1137C.
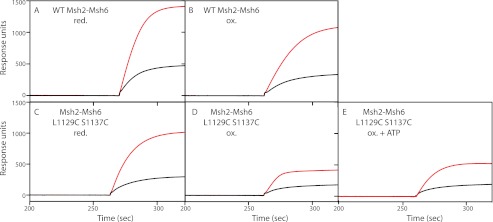
Oxidized Msh2-Msh6 L1129C/S1137C Is Defective for Mlh1-Pms1 Ternary Complex Formation
Surface plasmon resonance was also used to assess the ability of Msh2-Msh6 to assemble ternary complexes with Mlh1-Pms1 in the presence of ATP. In the presence of ATP, reduced or oxidized wild-type Msh2-Msh6 interacted with Mlh1-Pms1 and formed ternary complexes resulting in increased binding to the end-blocked mispaired substrate (Fig. 8, A and C, solid red line) compared with binding of Msh2-Msh6 alone (Fig. 8, A and C, dashed red line). Reduced Msh2-Msh6 L1129C/S1137C also mediated the formation of ternary complexes with Mlh1-Pms1, albeit to a lesser extent than wild-type Msh2-Msh6 (Fig. 8B). In contrast, oxidized Msh2-Msh6 L1129C/S1137C (Fig. 8D) and oxidized Msh2-Msh6 L1129C/S1137C that had been dialyzed overnight against buffer containing ATP (Fig. 8E) did not support the binding of Mlh1-Pms1. These results suggest that the ability of the Msh6 C-terminal region to switch between the two conformations is required for interaction between Msh2-Msh6 and Mlh1-Pms1.
FIGURE 8.
Msh2-Msh6 L1129C/S1137C can form a ternary complex with Mlh1-Pms1 under reducing conditions but not oxidizing conditions. Approximately 20 ng of mispair-containing DNA was conjugated to a flow cell of a streptavidin-coated Biacore SA chip. The dashed lines depict experiments in which 20 nm wild-type or mutant Msh2-Msh6, 30 nm LacI, and 250 μm were flowed over the chip. In the plus Mlh1-Pms1 experiments, which are depicted by the red solid line, the injection contained 20 nm Msh2-Msh6, 40 nm Mlh1-Pms1, 30 nm LacI, and 250 μm ATP. The small contribution of Mlh1-Pms1 alone binding the mispaired DNA was subtracted from the plus Mlh1-Pms1 experiments.
Oxidized Msh2-Msh6 L1129C/S1137C Has a Low Affinity for ATP
We previously demonstrated that ATP binding to both the Msh2 and Msh6 subunits as well as communication between the subunits is required for sliding clamp formation, and that ATP binding to only the Msh6 subunit is required for ternary complex formation (41). We used ATP cross-linking to determine whether the inability of oxidized Msh2-Msh6 L1129C/S1137C to form sliding clamps or ternary complexes with Mlh1-Pms1 might result from a reduced affinity for ATP. The absence of Mg2+ in the cross-linking reaction buffer prevented ATP hydrolysis but could have had a minor influence on the nucleotide binding characteristics of Msh2-Msh6 (28, 52). Reduced and oxidized wild-type Msh2-Msh6 had similar affinities for ATP. Consistent with the results of previous studies (28), 50% saturation of ATP (S0.5) in wild-type Msh6 for ATP was achieved at concentrations below 1 μm, and the S0.5 of Msh2 for ATP was between 20 and 50 μm (Fig. 9). In reduced Msh2-Msh6 L1129C/S1137C, the S0.5 of Msh2 and Msh6 for ATP were similar to each other and were between 20 and 50 μm, similar to the affinity of Msh2 for ATP in wild-type Msh2-Msh6. This indicates that in reduced Msh2-Msh6 L1129C/S1137C, the Msh6 subunit has a lower affinity for ATP relative to that in wild-type Msh2-Msh6; however, reduced Msh2-Msh6 L1129C/S1137C is likely proficient for sliding clamp formation and the formation of ternary complexes because the assays for these features were performed in buffer containing 250 μm ATP, a concentration significantly higher than the S0.5 for ATP binding to each subunit.
FIGURE 9.
Oxidized Msh2-Msh6 L1129C/S1137C has a reduced affinity for ATP in both the Msh2 and Msh6 ATP-binding sites. Wild-type or mutant Msh2-Msh6 was incubated with [γ-32P]ATP, cross-linked, and fractionated by SDS-PAGE.
In contrast, the affinities of both Msh2 and Msh6 for ATP were significantly reduced in oxidized Msh2-Msh6 L1129C/S1137C, with the S0.5 for ATP for both Msh2 and Msh6 being between 50 and 250 μm (Fig. 9). This result supports the possibility that the inability of oxidized mispair-bound Msh2-Msh6 L1129C/S1137C to form either sliding clamps or ternary complexes with Mlh1-Pms1 is due to the drastically reduced affinities of both the Msh2 and Msh6 subunits for ATP. Interestingly, both subunits, particularly Msh6 L1129C/S1137C, of the oxidized Msh2-Msh6 double mutant, which had been dialyzed in ATP overnight in the absence of Mg2+ and had undergone the ATP-induced conformational change as evidenced by the partial proteolysis pattern (Fig. 5), had even weaker affinities for ATP than those of oxidized Msh2-Msh6 L1129C/S1137C. This is most likely due to stable binding of unlabeled ATP, which forced the mutant to undergo the conformational change, competing with the radioactively labeled ATP in the cross-linking reaction. The severely reduced affinity of ATP-supplemented oxidized Msh2-Msh6 L1129C/S1137C for ATP is consistent with its inability to form the sliding clamp or ternary complex.
DISCUSSION
The Msh2-Msh6 complex appears to exist in at least two biochemically distinct forms that are modulated by ATP binding and hydrolysis. The nucleotide free and ADP bound forms are proficient for mispair binding (19, 28). ATP binding prevents Msh2-Msh6 from binding mispairs, but converts the mispair bound form to a sliding clamp and a form that recruits Mlh1-Pms1 (18–20, 24, 28, 41). These changes in biochemical properties are also mirrored by changes in partial proteolysis (36, 41) and are consistent with results from electron microscopy (30), deuterium exchange (42), analytical ultracentrifugation (38), and single-molecule FRET (53). Despite this, the requirement for conformational changes in the MutS family of proteins during MMR is not well understood (30, 38, 41, 42). To understand the role this conformational change plays in the activities of Msh2-Msh6 and MMR, we sought to introduce a reversible disulfide cross-link to prevent Msh2-Msh6 from transitioning between states.
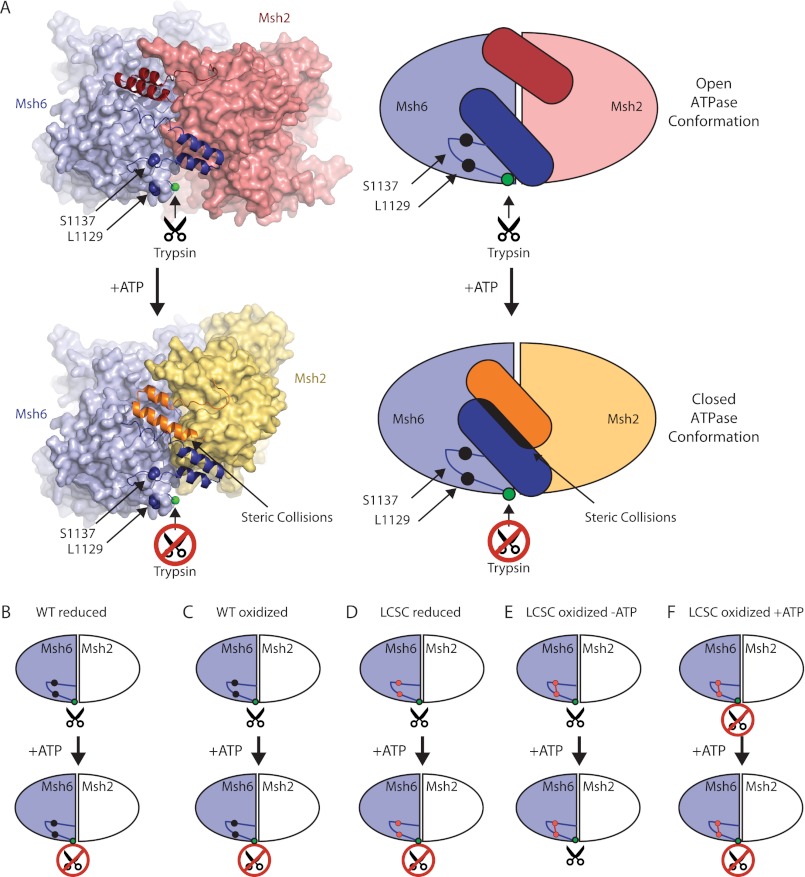
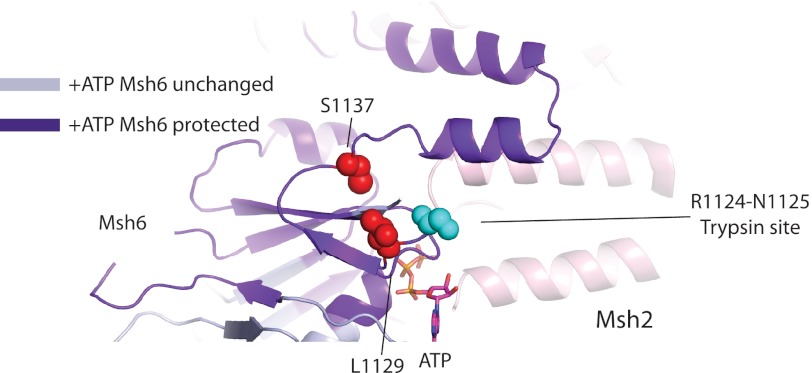
To model the effects of ATP binding on the ATPase domains of Msh2-Msh6, we compared the DNA-bound structures of MutS, Msh2-Msh6, and Msh2-Msh3, which are considered to be the open form of the ABC ATPases (2, 10, 39, 40, 54, 55), with the ATP-bound form of Rad50, which is considered to be the closed form of the ABC ATPases (43). This analysis indicated that the ATPase domains could readily switch between an open ATP-free conformation and a closed ATP-bound conformation, except for the final two ordered helices of both Msh2 and Msh6 in the DNA co-crystal structure (39). These helix pairs most likely move relative to the rest of the ATPase domain (Fig. 10A) due to steric collision with each other in the ATP-bound form as inferred from simple superposition of Msh2 and Msh6 onto Rad50 (Fig. 10A) or potentially by rigid separation of these helix pairs by a C-terminal dimerization domain equivalent to that of E. coli MutS (56) or human Msh2-Msh3 (40) that is disordered in the human Msh2-Msh6 crystal structure (39). The existence of this dimerization domain in S. cerevisiae Msh2-Msh6 and its requirement in MMR, as with Thermus aquaticus and E. coli MutS (56, 57), would explain why all of the Msh6 C-terminal residues, except for the last seven, are required for MMR (Fig. 2A). Conservation of this domain across MutS family members may be required to stabilize dimers so that the interface between the ATPase domains can change dynamically upon ATP binding. Regardless of the cause, movement of the final ordered helices of Msh2-Msh6 relative to the rest of the ATPase domain would move the loop containing Msh6 residues Leu1129 and Ser1137, and is consistent with ATP binding causing increased protection from deuterium exchange (Fig. 11) (42) and protection of the Arg1124-Asn1125 trypsin site from cleavage (Fig. 1).
FIGURE 10.
Model of the open to closed transition of Msh2-Msh6 caused by ATP binding. A, diagram of human Msh2-Msh6 with the two C-terminal α helices in ribbons and the rest of the protein in spheres and a representative schematic. The ATP-bound conformation of human Msh2-Msh6 was modeled by individually superimposing the human Msh2 and Msh6 subunits (PDB code 2O8B) on the ATP-bound form of Pyrococcus furiosus Rad50 (PDB code 1F2U (43)) using Sequoia (60). Views were chosen so that the Msh6 orientation is fixed. Schematics of reduced wild-type Msh2-Msh6 (B), oxidized wild-type Msh2-Msh6 (C), reduced Msh2-Msh6 L1129C/S1137C (D), oxidized Msh2-Msh6 L1129C/S1137C (E), and ATP-supplemented oxidized Msh2-Msh6 L1129C/S1137C (F).
FIGURE 11.
Ribbon diagram of Homo sapiens Msh2-Msh6 (PDB ID code 2O8B) with S. cerevisiae residues L1129C and S1137C and the Arg1124-Asn1125 trypsin cleavage site in spheres, ATP in sticks, residues protected from deuterium exchange by ATP binding depicted in purple, and residues unchanged upon ATP binding depicted in gray (42).
Introduction of double cysteine amino acid substitutions, L1129C and S1137C, into the loop of Msh6 preceding the final ordered helices generated a protein that formed a disulfide cross-link upon oxidation (Fig. 4). Genetically, the double cysteine mutation reduced MMR proficiency in vivo albeit not to the same extent as caused by deleting MSH6 (Fig. 3A, Table 2). One potential reason why an MSH6 deletion causes a higher mutation rate is that the cytoplasm of S. cerevisiae is reducing (58) and hence disulfide bond formation may not go to completion in vivo. However, under reducing conditions in vitro, 31% of mutant protein contained a L1192C/S1137C disulfide bond (Fig. 4). Taken together, this suggests that a sufficient fraction of protein containing the disulfide bond can form in vivo to produce the elevated mutation rate observed in vivo.
The reduced Msh2-Msh6 L1129C/S1137C behaved like wild-type in the accessibility of the Arg1124-Asn1125 trypsin site in the open and closed conformations (Figs. 5 and 10D), the formation of the sliding clamp (Fig. 6), and recruitment of Mlh1-Pms1 (Fig. 8). The greatest difference between the reduced mutant and wild-type proteins was that the mutant Msh6 had a reduced affinity for ATP, which was equivalent to Msh2 in wild-type Msh2-Msh6 and Msh2-Msh6 L1129C/S1137C. Sliding clamp formation and recruitment of Mlh1-Pms1 differ in their ATP requirements in that recruitment of Mlh1-Pms1 only requires ATP binding to the high affinity Msh6 site, whereas sliding clamp formation requires ATP binding to both the Msh2 and Msh6 sites (41); however, the concentration of ATP in the experiment, 250 μm, was sufficient to saturate the ATP binding sites of the Msh2 and Msh6 subunits in this mutant explaining how it can be proficient for sliding clamp formation and recruitment of Mlh1-Pms1. This contrasts with other mutants, such as Msh2-R730C-Msh6 and Msh2-Msh6-R1024C, which were unable to form sliding clamps or bind Mlh1-Pms1 even when the ATP concentration was above the measured S0.5 values for each subunit; Msh2-R730C-Msh6 and Msh2-Msh6-R1024C most likely have additional defects in communication between the ATP-binding sites (41).
The oxidized Msh2-Msh6 L1129C/S1137C, however, appeared to be defective for normal rapid rates of ATP binding as evidenced by the generation of the 75-kDa band in the presence of ATPγS (Figs. 5 and 10E), its ability to bind mispairs (Fig. 7), its failure to form a sliding clamp or recruit Mlh1-Pms1 (Figs. 6–8), and its reduced affinity for ATP in both subunits (Fig. 9). Although these effects can be attributed to lack of ATP binding by Msh6, the L1129C/S1137C disulfide bond is not at the nucleotide-binding site; it is located in a region undergoing ATP-induced changes according to deuterium exchange data (42). Thus, inhibition of rapid ATP binding is more likely due to disruption of ATP-induced conformational changes that generate the high affinity ATP-binding site, rather than disruption of the nucleotide-binding site per se. The fact that extensive dialysis of oxidized Msh2-Msh6 L1129C/S1137C against buffer containing ATP without Mg2+ converted it to a state in which trypsin was unable to generate the 75-kDa fragment (Figs. 5 and 10F) is consistent with a slowed ATP-induced transition and is at odds with models in which the disulfide bond directly disrupts the nucleotide-binding site. Thus, the formation of the disulfide bond appears to have generated an Msh2-Msh6 complex in which the ATP-induced conformational changes occur on the order of hours.
Supplementing oxidized Msh2-Msh6 L1129C/S1137C with ATP reduced the affinity of both its subunits for ATP relative to oxidized Msh2-Msh6 L1129C/S1137C. The unlabeled ATP from the dialysis most likely competed with the added labeled ATP in the UV cross-linking experiment (Fig. 9). The ATP-supplemented oxidized Msh6 L1129C/S1137C subunit appeared to retain more unlabeled ATP than Msh2, resulting in less cross-linked labeled ATP. This result suggests that the C-terminal region of Msh6 affects ATP binding to Msh2, consistent with previous results (28, 41, 52). Interestingly, this ATP-bound form of oxidized Msh2-Msh6 L1129C/S1137C was capable of binding mispaired bases (Fig. 7), but did not form the sliding clamp or recruit Mlh1-Pms1 (Figs. 6 and 8). This is most likely because the bound ATP was hydrolyzed to ADP due to the presence of Mg2+ in the Biacore running buffer, which converted it to a mispair-binding proficient form (19, 28), but the slow ATP binding prevented either sliding clamp formation or Mlh1-Pms1 binding.
The fact that in vitro oxidized Msh2-Msh6 L1129C/S1137C is in the open conformation but can be slowly converted to the closed conformation (Figs. 5 and 10) indicates that the disulfide bond slows ATP binding to Msh2 and Msh6, and hence the dynamic interconversion between the open and closed forms that would be predicted to occur during MMR. Therefore the C-terminal conformational change in Msh6 plays a role in ATP processing, specifically ATP hydrolysis or ADP exchange for ATP. This, in turn, disrupts the formation of both Msh2-Msh6 sliding clamps and ternary complexes between Msh2-Msh6 and Mlh1-Pms1 and results in an MMR defect.
Acknowledgments
We thank Kathleen Matthews for the generous gift of LacI protein. We thank Jason Liang and Huilin Zhou for their assistance in analyzing the disulfide bond formation by mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Grants GM50006 and CA092584.
- MMR
- mismatch repair
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1. Schofield M. J., Hsieh P. (2003) DNA mismatch repair. Molecular mechanisms and biological function. Annu. Rev. Microbiol. 57, 579–608 [DOI] [PubMed] [Google Scholar]
- 2. Lamers M. H., Perrakis A., Enzlin J. H., Winterwerp H. H., de Wind N., Sixma T. K. (2000) The crystal structure of DNA mismatch repair protein MutS binding to a GXT mismatch. Nature 407, 711–717 [DOI] [PubMed] [Google Scholar]
- 3. Marti T. M., Kunz C., Fleck O. (2002) DNA mismatch repair and mutation avoidance pathways. J. Cell. Physiol. 191, 28–41 [DOI] [PubMed] [Google Scholar]
- 4. Iyer R. R., Pluciennik A., Burdett V., Modrich P. L. (2006) DNA mismatch repair. Functions and mechanisms. Chem. Rev. 106, 302–323 [DOI] [PubMed] [Google Scholar]
- 5. Li G. M. (2008) Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 [DOI] [PubMed] [Google Scholar]
- 6. Lynch H. T., de la Chapelle A. (1999) Genetic susceptibility to non-polyposis colorectal cancer. J. Med. Genet. 36, 801–818 [PMC free article] [PubMed] [Google Scholar]
- 7. Wheeler J. M., Bodmer W. F., Mortensen N. J. (2000) DNA mismatch repair genes and colorectal cancer. Gut 47, 148–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lynch H. T., de la Chapelle A. (2003) Hereditary colorectal cancer. N. Engl. J. Med. 348, 919–932 [DOI] [PubMed] [Google Scholar]
- 9. Peltomäki P. (2003) Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 21, 1174–1179 [DOI] [PubMed] [Google Scholar]
- 10. Obmolova G., Ban C., Hsieh P., Yang W. (2000) Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407, 703–710 [DOI] [PubMed] [Google Scholar]
- 11. Su S. S., Modrich P. (1986) Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc. Natl. Acad. Sci. U.S.A. 83, 5057–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiricny J. (2006) MutLα. At the cutting edge of mismatch repair. Cell 126, 239–241 [DOI] [PubMed] [Google Scholar]
- 13. Hopfner K. P., Tainer J. A. (2000) DNA mismatch repair. The hands of a genome guardian. Structure 8, R237–241 [DOI] [PubMed] [Google Scholar]
- 14. Marsischky G. T., Kolodner R. D. (1999) Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J. Biol. Chem. 274, 26668–26682 [DOI] [PubMed] [Google Scholar]
- 15. Marsischky G. T., Filosi N., Kane M. F., Kolodner R. (1996) Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10, 407–420 [DOI] [PubMed] [Google Scholar]
- 16. Acharya S., Wilson T., Gradia S., Kane M. F., Guerrette S., Marsischky G. T., Kolodner R., Fishel R. (1996) hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl. Acad. Sci. U.S.A. 93, 13629–13634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Genschel J., Littman S. J., Drummond J. T., Modrich P. (1998) Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J. Biol. Chem. 273, 19895–19901 [DOI] [PubMed] [Google Scholar]
- 18. Selmane T., Schofield M. J., Nayak S., Du C., Hsieh P. (2003) Formation of a DNA mismatch repair complex mediated by ATP. J. Mol. Biol. 334, 949–965 [DOI] [PubMed] [Google Scholar]
- 19. Acharya S., Foster P. L., Brooks P., Fishel R. (2003) The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell 12, 233–246 [DOI] [PubMed] [Google Scholar]
- 20. Blackwell L. J., Wang S., Modrich P. (2001) DNA chain length dependence of formation and dynamics of hMutSα·hMutLα·heteroduplex complexes. J. Biol. Chem. 276, 33233–33240 [DOI] [PubMed] [Google Scholar]
- 21. Flores-Rozas H., Kolodner R. D. (1998) The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. U.S.A. 95, 12404–12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grilley M., Welsh K. M., Su S. S., Modrich P. (1989) Isolation and characterization of the Escherichia coli mutL gene product. J. Biol. Chem. 264, 1000–1004 [PubMed] [Google Scholar]
- 23. Hall M. C., Matson S. W. (1999) The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem. 274, 1306–1312 [DOI] [PubMed] [Google Scholar]
- 24. Mendillo M. L., Mazur D. J., Kolodner R. D. (2005) Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J. Biol. Chem. 280, 22245–22257 [DOI] [PubMed] [Google Scholar]
- 25. Welsh K. M., Lu A. L., Clark S., Modrich P. (1987) Isolation and characterization of the Escherichia coli mutH gene product. J. Biol. Chem. 262, 15624–15629 [PubMed] [Google Scholar]
- 26. Räschle M., Marra G., Nyström-Lahti M., Schär P., Jiricny J. (1999) Identification of hMutLβ, a heterodimer of hMLH1 and hPMS1. J. Biol. Chem. 274, 32368–32375 [DOI] [PubMed] [Google Scholar]
- 27. Haber L. T., Walker G. C. (1991) Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 10, 2707–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mazur D. J., Mendillo M. L., Kolodner R. D. (2006) Inhibition of Msh6 ATPase activity by mispaired DNA induces a Msh2(ATP)-Msh6(ATP) state capable of hydrolysis-independent movement along DNA. Mol. Cell 22, 39–49 [DOI] [PubMed] [Google Scholar]
- 29. Gradia S., Acharya S., Fishel R. (1997) The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell 91, 995–1005 [DOI] [PubMed] [Google Scholar]
- 30. Gradia S., Subramanian D., Wilson T., Acharya S., Makhov A., Griffith J., Fishel R. (1999) hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell 3, 255–261 [DOI] [PubMed] [Google Scholar]
- 31. Allen D. J., Makhov A., Grilley M., Taylor J., Thresher R., Modrich P., Griffith J. D. (1997) MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 16, 4467–4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blackwell L. J., Martik D., Bjornson K. P., Bjornson E. S., Modrich P. (1998) Nucleotide-promoted release of hMutSα from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 273, 32055–32062 [DOI] [PubMed] [Google Scholar]
- 33. Junop M. S., Obmolova G., Rausch K., Hsieh P., Yang W. (2001) Composite active site of an ABC ATPase. MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell 7, 1–12 [DOI] [PubMed] [Google Scholar]
- 34. Alani E., Sokolsky T., Studamire B., Miret J. J., Lahue R. S. (1997) Genetic and biochemical analysis of Msh2p-Msh6p. Role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol. Cell. Biol. 17, 2436–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Das Gupta R., Kolodner R. D. (2000) Novel dominant mutations in Saccharomyces cerevisiae MSH6. Nat. Genet. 24, 53–56 [DOI] [PubMed] [Google Scholar]
- 36. Studamire B., Quach T., Alani E. (1998) Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol. Cell. Biol. 18, 7590–7601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson T., Guerrette S., Fishel R. (1999) Dissociation of mismatch recognition and ATPase activity by hMSH2-hMSH3. J. Biol. Chem. 274, 21659–21664 [DOI] [PubMed] [Google Scholar]
- 38. Lamers M. H., Georgijevic D., Lebbink J. H., Winterwerp H. H., Agianian B., de Wind N., Sixma T. K. (2004) ATP increases the affinity between MutS ATPase domains. Implications for ATP hydrolysis and conformational changes. J. Biol. Chem. 279, 43879–43885 [DOI] [PubMed] [Google Scholar]
- 39. Warren J. J., Pohlhaus T. J., Changela A., Iyer R. R., Modrich P. L., Beese L. S. (2007) Structure of the human MutSα DNA lesion recognition complex. Mol. Cell 26, 579–592 [DOI] [PubMed] [Google Scholar]
- 40. Gupta S., Gellert M., Yang W. (2012) Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 19, 72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hargreaves V. V., Shell S. S., Mazur D. J., Hess M. T., Kolodner R. D. (2010) Interaction between the Msh2 and Msh6 nucleotide-binding sites in the Saccharomyces cerevisiae Msh2-Msh6 complex. J. Biol. Chem. 285, 9301–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mendillo M. L., Putnam C. D., Mo A. O., Jamison J. W., Li S., Woods V. L., Jr., Kolodner R. D. (2010) Probing DNA- and ATP-mediated conformational changes in the MutS family of mispair recognition proteins using deuterium exchange mass spectrometry. J. Biol. Chem. 285, 13170–13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., Tainer J. A. (2000) Structural biology of Rad50 ATPase. ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 [DOI] [PubMed] [Google Scholar]
- 44. Mendillo M. L., Hargreaves V. V., Jamison J. W., Mo A. O., Li S., Putnam C. D., Woods V. L., Jr., Kolodner R. D. (2009) A conserved MutS homolog connector domain interface interacts with MutL homologs. Proc. Natl. Acad. Sci. U.S.A. 106, 22223–22228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Winkler I., Marx A. D., Lariviere D., Heinze R. J., Cristovao M., Reumer A., Curth U., Sixma T. K., Friedhoff P. (2011) Chemical trapping of the dynamic MutS-MutL complex formed in DNA mismatch repair in Escherichia coli. J. Biol. Chem. 286, 17326–17337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Amin N. S., Nguyen M. N., Oh S., Kolodner R. D. (2001) exo1-Dependent mutator mutations. Model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 21, 5142–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Antony E., Hingorani M. M. (2003) Mismatch recognition-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry 42, 7682–7693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang M. E., Kolodner R. D. (2005) A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol. Cell 17, 709–720 [DOI] [PubMed] [Google Scholar]
- 49. Shell S. S., Putnam C. D., Kolodner R. D. (2007) Chimeric Saccharomyces cerevisiae Msh6 protein with an Msh3 mispair-binding domain combines properties of both proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 10956–10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hess M. T., Gupta R. D., Kolodner R. D. (2002) Dominant Saccharomyces cerevisiae msh6 mutations cause increased mispair binding and decreased dissociation from mispairs by Msh2-Msh6 in the presence of ATP. J. Biol. Chem. 277, 25545–25553 [DOI] [PubMed] [Google Scholar]
- 51. Lundgren D. H., Martinez H., Wright M. E., Han D. K. (2009) Current Protocols in Bioinformatics (Baxevanis A. D., Pearson W. R., Stein L. D., Stormo G. D., Yeates J. R., III, eds) Chapter 13, Unit 13.3 John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 52. Heinen C. D., Cyr J. L., Cook C., Punja N., Sakato M., Forties R. A., Lopez J. M., Hingorani M. M., Fishel R. (2011) Human MSH2 (hMSH2) protein controls ATP processing by hMSH2-hMSH6. J. Biol. Chem. 286, 40287–40295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Qiu R., DeRocco V. C., Harris C., Sharma A., Hingorani M. M., Erie D. A., Weninger K. R. (2012) Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. EMBO J. 31, 2528–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Alani E., Lee J. Y., Schofield M. J., Kijas A. W., Hsieh P., Yang W. (2003) Crystal structure and biochemical analysis of the MutS·ADP·beryllium fluoride complex suggests a conserved mechanism for ATP interactions in mismatch repair. J. Biol. Chem. 278, 16088–16094 [DOI] [PubMed] [Google Scholar]
- 55. Natrajan G., Lamers M. H., Enzlin J. H., Winterwerp H. H., Perrakis A., Sixma T. K. (2003) Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches. A common recognition mode for diverse substrates. Nucleic Acids Res. 31, 4814–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mendillo M. L., Putnam C. D., Kolodner R. D. (2007) Escherichia coli MutS tetramerization domain structure reveals that stable dimers but not tetramers are essential for DNA mismatch repair in vivo. J. Biol. Chem. 282, 16345–16354 [DOI] [PubMed] [Google Scholar]
- 57. Biswas I., Obmolova G., Takahashi M., Herr A., Newman M. A., Yang W., Hsieh P. (2001) Disruption of the helix-u-turn-helix motif of MutS protein. Loss of subunit dimerization, mismatch binding and ATP hydrolysis. J. Mol. Biol. 305, 805–816 [DOI] [PubMed] [Google Scholar]
- 58. López-Mirabal H. R., Winther J. R. (2008) Redox characteristics of the eukaryotic cytosol. Biochim. Biophys. Acta 1783, 629–640 [DOI] [PubMed] [Google Scholar]
- 59. DeLano W. (2002) The PyMol Molecular Graphics System, DeLano Scientific, South San Francisco, CA [Google Scholar]
- 60. Bruns C. M., Hubatsch I., Ridderström M., Mannervik B., Tainer J. A. (1999) Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products. J. Mol. Biol. 288, 427–439 [DOI] [PubMed] [Google Scholar]