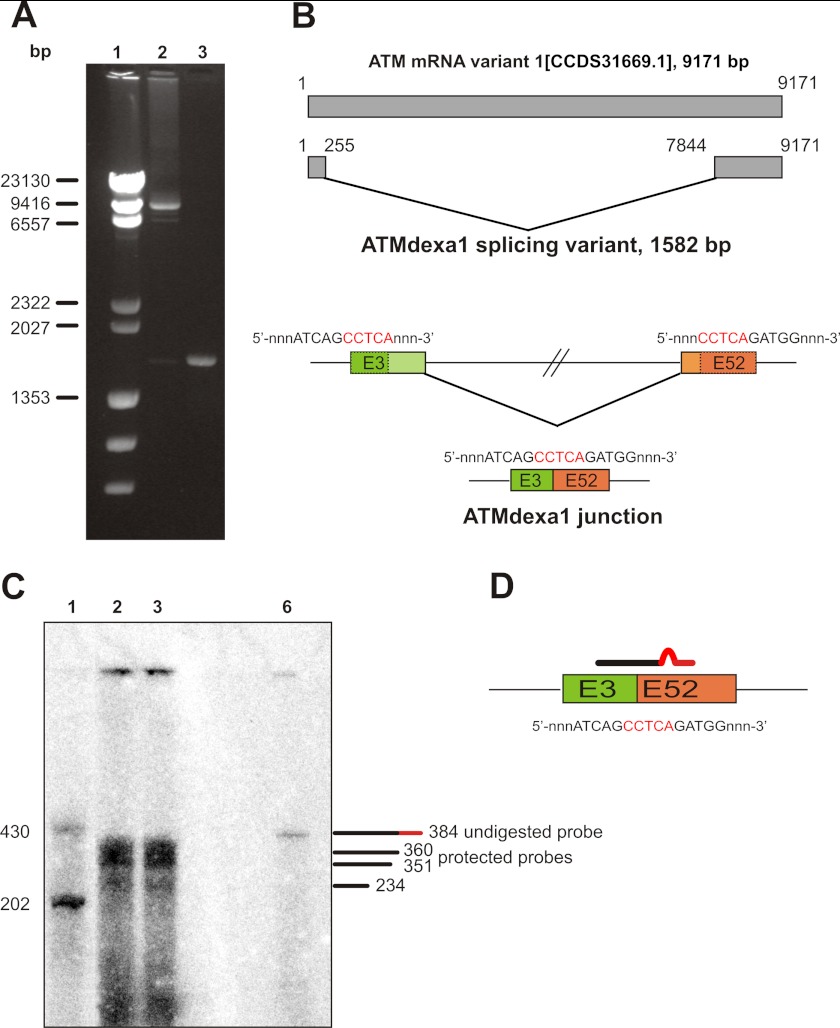
FIGURE 2.
Identification of an alternative ATM transcript induced by dexa. A, RT-PCR on AT lymphoblastoid cell lines. The entire CDS of ATM gene was amplified by RT-PCR on total cDNA templates obtained from AT lymphoblastoid cell lines treated or not with dexa. The full-length CDS was amplified in control cells, whereas a shorter band, lacking the central portion of ATM CDS, was identified in treated cells. Lane 1, λ HindIII DNA ladder; lane 2, untreated AT129RM; lane 3, treated AT129RM. B, scheme representing the alternative ATM splicing variant induced by dexa. Sequencing of the deleted band identified in AT LCLs reveals a noncanonical splicing event in the ATM gene, mediated by a short direct repeat (in red), which leads to the joining of exon 3 with exon 52. The resulting cDNA carries the first three exons and the last 11 exons of native CDS and lacks internal ones. C, S1 nuclease protection assay on AT lymphoblastoid cell lines. To prove the real attendance of SDR-directed spliced ATM mRNA (ATMdexa1), a S1 nuclease protection assay was performed. Total RNA from AT lymphoblastoid cell lines receiving 100 nm dexa was probed with a specific probe complementary to the identified junction and hybridization mixture subjected to S1 digestion. Protection of the probe demonstrates the presence of ATMdexa1 mRNA in both AT lines. Lane 1, ssDNA ladder; lane 2, AT28RM; lane 3, AT129RM; lane 6, undigested probe. D, exemplification of the probe designed for S1 protection assay overlapping the ATMdexa1 junction.