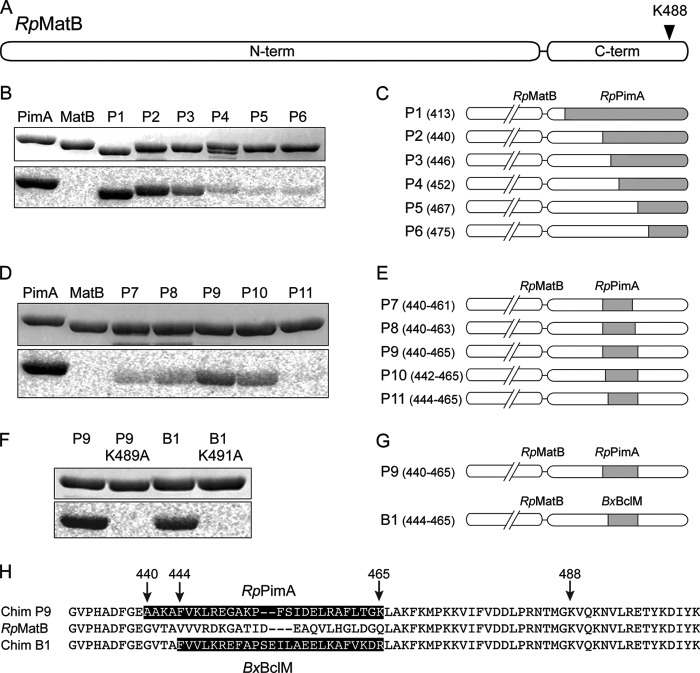
FIGURE 2.
Construction of RpMatB chimeric proteins that can be acetylated. A, scheme of RpMatB N- and C-terminal domains drawn to scale. Arrowhead indicates location of conserved lysine residue Lys-488. B, acetylation of six RpMatB-RpPimA chimeras by RpPat using [1-14C]acetyl-CoA. Top panel shows SDS-polyacrylamide gel, and bottom panel shows the phosphorimage of the same gel. C, schematic representation of the six RpMatB-RpPimA chimeras P1–P6, in which a section of the C-terminal domain of RpMatB was replaced with the corresponding sequence from RpPimA (gray). C-terminal domains are drawn to match scale of A. Numbers in parentheses indicate the residue of RpMatB at which the fusion begins (i.e. first residue replaced by PimA). D, acetylation by RpPat of RpMatB chimeras containing an internal fragment of RpPimA. E, schematic representation of RpMatB-RpPimA chimeras P7–P11, containing an internal fragment of RpPimA (gray). Numbers in parentheses indicate residues of RpMatB that were replaced by the corresponding sequence of RpPimA. F, acetylation by RpPat of the RpMatB-BxBclM chimera B1. Changing the conserved lysine residue (Lys-488 in wild-type RpMatB numbering) in either chimera resulted in no acetylation by RpPat. G, schematic representation of the RpMatB-RpPimA chimera P9 and RpMatB-BxBclM chimera B1 that were the best identified substrates of RpPat. H, alignment of the C-terminal ends of RpMatB and the RpMatB-RpPimA P9 and RpMatB-BxBclM B1 chimeras, starting at Gly-431. The sequences derived from RpPimA and BxBclM are shaded, and RpMatB residue numbers are indicated with arrows.