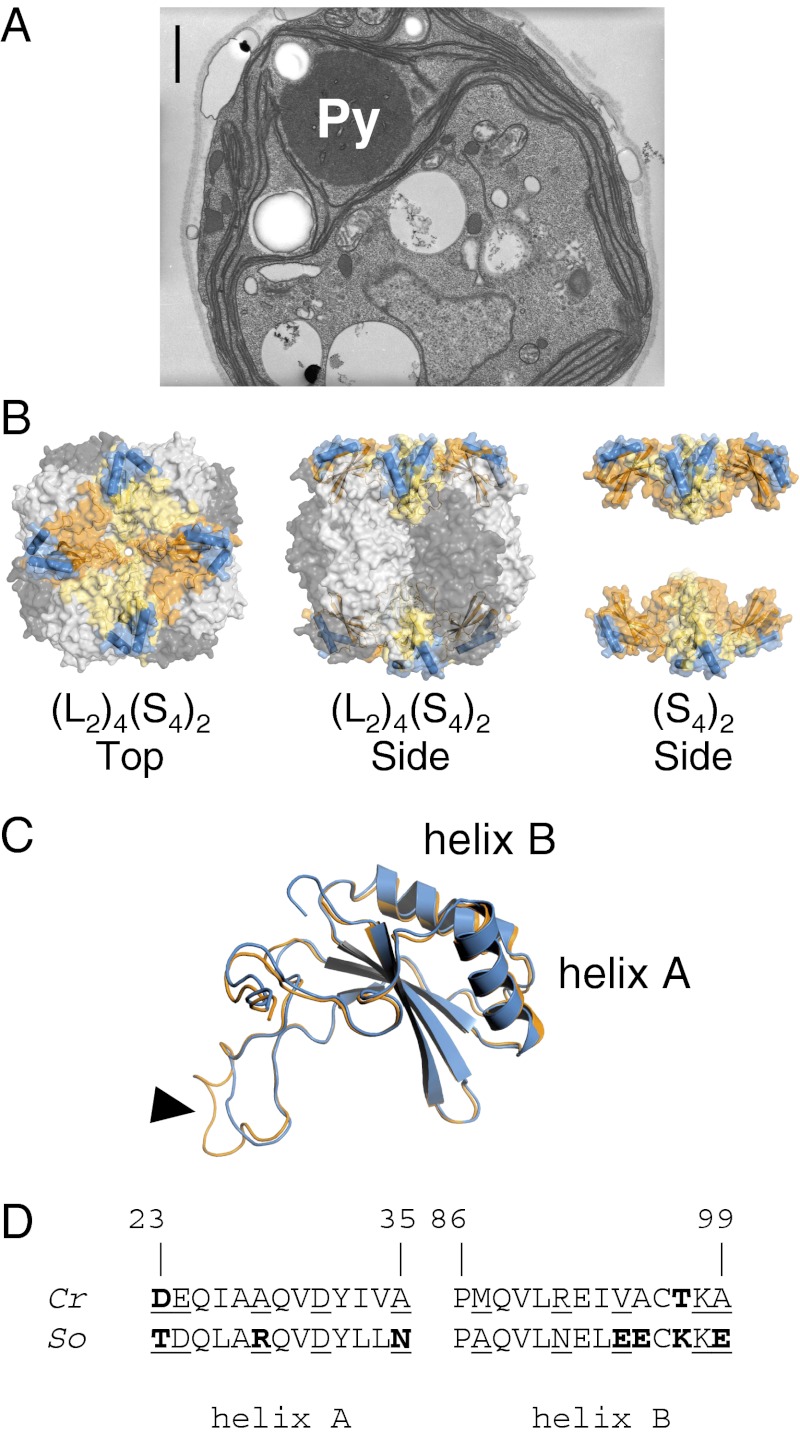
Fig. 1.
(A) Electron micrograph of a wild-type Chlamydomonas cell, grown under air-level CO2. Rubisco is packaged into a single conspicuous pyrenoid (Py). (Scale bar, 1 µm.) (B) Structure of the hexadecameric Rubisco form I of Chlamydomonas. The catalytic core is composed of four antiparallel LSU dimers (dark/light gray), stabilized by eight SSUs. Each SSU (orange/yellow) has two solvent-exposed α-helices (blue) that contribute to one-third of the surface-exposed SSU residues. (C) Despite 50% difference in amino acid composition, folding of Chlamydomonas (orange) and spinach (blue) SSU is highly conserved, as shown by the near-perfect overlay. The algal and higher plant SSUs differ primarily by the length of the βA–βB loop (arrowhead), which extends into the solvent channel of the holoenzyme. (D) Comparison of the amino acid composition of the Chlamydomonas reinhardtii (Cr) and Spinacia oleracea (So) SSU α-helices (22, 23). Numbering is relative to the Chlamydomonas sequence (24). The helices in spinach are distinctly more hydrophilic than in Chlamydomonas. Hydrophilic/polar residues differing between the two species are highlighted in bold; solvent-exposed residues are underlined.