Fig. P1.
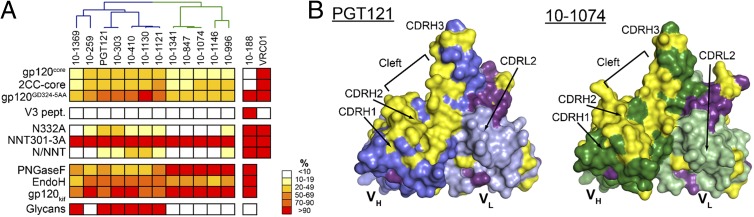
(A) Heat map (expressed as percentage of binding to unmodified gp120) summarizing the binding of PGT121-like and 10-1074–like antibodies to antigens. Darker colors represent stronger binding; white represents no detectable binding. A dendrogram above the antibody names divides them into PGT121-like (blue lines) and 10-1074–like (green lines) antibodies. Antibodies against the gp120 V3 loop (10-188) and CD4-binding site (VRC01) are controls. For V3 loop peptide-binding assays (V3 pept.) and glycan microarrays (Glycans), red indicates binding and white indicates no binding. PNGase F and Endo H are glycosidase enzymes that can cleave complex-type and high-mannose glycans (PNGase F) or high-mannose glycans (Endo H). N332A indicates the substitution of asparagine at gp120 position 332 to alanine. (B) Surface representation of PGT121 (blue) and 10-1074 (green) variable domain structures showing the location of differences (yellow) and somatic mutations in common (purple).