Abstract
Background
Melatonin receptor 1B (MTNR1B) belongs to the seven-transmembrane G protein-coupled receptor superfamily involved in insulin secretion, which has attracted considerable attention as a candidate gene for type 2 diabetes (T2D) since it was first identified as a loci associated with fasting plasma glucose level through genome wide association approach. The relationship between MTNR1B and T2D has been reported in various ethnic groups. The aim of this study was to consolidate and summarize published data on the potential of MTNR1B polymorphisms in T2D risk prediction.
Methods
PubMed, EMBASE, ISI web of science and the CNKI databases were systematically searched to identify relevant studies. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated. Heterogeneity and publication bias were also tested.
Results
A total of 23 studies involving 172,963 subjects for two common polymorphisms (rs10830963, rs1387153) on MTNR1B were included. An overall random effects per-allele OR of 1.05 (95% CI: 1.02–1.08; P<10−4) and 1.04 (95% CI: 0.98–1.10; P = 0.20) were found for the two variants respectively. Similar results were also observed using dominant or recessive genetic model. There was strong evidence of heterogeneity, which largely disappeared after stratification by ethnicity. Significant results were found in Caucasians when stratified by ethnicity; while no significant associations were observed in East Asians and South Asians. Besides, we found that the rs10830963 polymorphism is a risk factor associated with increased impaired glucose regulation susceptibility.
Conclusions
This meta-analysis demonstrated that the rs10830963 polymorphism is a risk factor for developing impaired glucose regulation and T2D.
Introduction
Glucose homeostasis in healthy individuals is tightly controlled through a complex pathway of regulatory mechanisms involving multiple organs and tissues. Disruption of normal glucose homeostasis and substantial elevations of fasting glucose are hallmarks of type 2 diabetes (T2D) and typically result from sustained reduction in pancreatic beta-cell function and insulin secretion. However, there is substantial variation in fasting glucose levels even within healthy, non-diabetic populations. Approximately one-third of this variation is genetic [1], but little of this heritability has been explained.
Recent genome-wide association studies (GWAS) and meta-analysis have identified genes contributing to the variation of fasting plasma glucose (FPG) levels in populations of European origin [2]–[6]. Two common variants (rs10830963, rs1387153) in the melatonin receptor 1B (MTNR1B) were shown to have moderate effects on FPG levels in nondiabetic individuals with an increased risk for T2D [2], [4]. MTNR1B is the receptor of melatonin which inhibits insulin secretion through its effect on the formation of cGMP [7], [8]. Knock-out mice of these genes demonstrated significantly lower fasting glucose levels [9], [10]. Over the past few years, considerable efforts have been devoted to exploring the relationships between the MTNR1B polymorphisms and T2D. Genetic association studies can be problematic to reproduce due to multiple hypothesis testing, population stratification, source of controls, publication bias, and phenotypic heterogeneity. In addition, with the increased studies in recent years among Asian, and other populations, there is a need to reconcile these data. Therefore, we performed a meta-analysis of the published to establish a comprehensive picture of the relationship between MTNR1B and risk of T2D as well as to quantify the between-study heterogeneity and potential bias.
Materials and Methods
Literature search strategy
Genetic association studies published before the end of May 2012 on T2D and polymorphisms in the MTNR1B gene were identified through a search of PubMed, Web of Science, EMBASE and CNKI (Chinese National Knowledge Infrastructure). Search term were keywords relating to the relevant gene (e.g. ‘melatonin receptor 1B’ or ‘MTNR1B’) in combination with words related to T2D (e.g. ‘Type 2 diabetes’ or ‘Type 2 diabetes mellitus’ or ‘non-insulin dependent diabetes mellitus’). Furthermore, reference lists of main reports and review articles were also reviewed by a manual search to identify additional relevant publications.
Eligible studies and data extraction
The included studies have to meet the following criteria: (1) original papers containing independent data which have been published in peer-reviewed journal, (2) identification of T2D patients according to the World Health Organization criteria, American Diabetes Association criteria, or other standard criteria, (3) genotype distribution information or odds ratio (OR) with its 95% confidence interval (CI) and P-value;, (4) case–control or cohort studies. The major reasons for exclusion of studies were (1) overlapping data and (2) case-only studies, family based studies, and review articles.
Data extraction was performed independently by two reviewers and differences were resolved by further discussion among all authors. For each included study, the following information was extracted from each report according to a fixed protocol: first author, publication year, definition and numbers of cases and controls, diagnostic criterion, impaired glucose regulation (IGR) status (impaired fasting glucose and/or impaired glucose tolerance), frequency of genotypes, age, sex, body mass index (BMI), Hardy–Weinberg equilibrium status, ethnicity and genotyping method.
Statistical methods
Deviation from Hardy–Weinberg equilibrium for controls was examined by χ2 tests. Odds ratio (OR) with 95% confidence intervals (CIs) was used to assess the strength of association between the MTNR1B gene polymorphism and T2D risk. The per-allele OR of the risk allele was compared between cases and controls. Then, we examined the association between risk genotype of these polymorphisms and T2D susceptibility using dominant and recessive genetic models. Heterogeneity across individual studies was calculated using the Cochran chi-square Q test followed by subsidiary analysis or by random-effects regression models with restricted maximum likelihood estimation [11]–[13]. Random-effects and fixed-effect summary measures were calculated as inverse variance-weighted average of the log OR. The results of random-effects summary were reported in the text because it takes into account the variation between studies. The 95% CIs were constructed using Woolf's method [14]. The significance of the overall OR was determined by the Z-test. Sample size (No. cases ≥1000 or <1000) and ethnicity were prespecified as characteristics for assessment of heterogeneity. Ethnic group was defined as Caucasian (i.e., people of European origin), East Asian (e.g., Chinese, Japanese, and Korean), and South Asian (e.g., Indian, and Pakistani). In addition, sample size, ethnicity, gender distribution in cases and controls, genotyping method, mean age and BMI of cases and controls were analyzed as covariates in meta-regression.
Sensitivity analyses were performed to assess the stability of the results, namely, a single study in the meta-analysis was deleted each time to reflect the influence of the individual data set to the overall OR. Publication bias was assessed using Egger's test [15] and Begg's funnel plots [16]. All P values are two-sided at the P = 0.05 level. All analyses were conducted using the STATA 10.0 (STATA Corporation, College Station, TX).
Results
Characteristics of studies
The combined search yielded 72 references. Forty-nine articles were excluded because they clearly did not meet the criteria or overlapping references. Finally, a total of 23 studies were retrieved based on the search criteria for T2D susceptibility related to the MTNR1B polymorphisms [2], [4], [17]–[37] (Figure S1). The main study characteristics were summarized in Table 1. There are 16 studies with 51, 552 T2D cases and 92, 618 controls concerning rs10830963 and 7 studies with 14, 874 T2D cases and 17, 703 controls concerning rs1387153. These two polymorphisms were found to occur in frequencies consistent with Hardy-Weinberg equilibrium in the control populations of the vast majority of the published studies. Of the cases, 63% were Caucasians, 31% were East Asians, and 6% were South Asians.
Table 1. Characteristics of the studies included in the meta-analysis.
| Study | Year | Ethnicity | Cases | Controls | Polymorphism | No. of case | No. of control | Genotyping method |
| Staiger [17] | 2008 | German | IFG, IGT confirmed by OGTT | Normal glucose tolerant | rs10830963 | 287 IFG, 275 IGT | 1139 | TaqMan |
| Sparsø [18] | 2009 | European | T2D, IFG, IGT per WHO criteria | Normal fasting glycaemia; Normal glucose tolerant | rs10830963, rs1387153 | 1948 T2D, 686 IFG, 665 IGT | 4905 | Taqman |
| Rönn [19] | 2009 | Chinese | T2D per WHO criteria | Normoglycaemic | rs10830963 | 1165 T2D | 1105 | iPLEX |
| Prokopenko [2] | 2009 | European | T2D patients | Non-diabetic | rs10830963 | 18236 T2D | 64353 | Taqman, iPLEX, Illumina chip, Affymetrix chip |
| Bouatia-Naji [4] | 2009 | French, Danish | T2D per WHO criteria | Normal fasting glycaemia; Normal glucose tolerant | rs1387153 | 6332 T2D | 9132 | Illumina chip |
| Reiling [20] | 2009 | Dutch | T2D per WHO criteria | Normal glucose tolerant | rs10830963 | 2537 T2D | 1990 | Taqman |
| Tam [21] | 2010 | Chinese | T2D per WHO criteria | Normal fasting glucose | rs10830963 | 1342 T2D | 1644 | MassARRAY |
| Liu [22] | 2010 | Chinese | T2D per WHO criteria | Normal fasting glucose | rs10830963 | 424 T2D | 1908 | SNPstream |
| Simonis-Bik [23] | 2010 | European | IGT per WHO criteria | Normal glucose tolerant | rs10830963 | 158 IGT | 177 | Taqman |
| Xu [24] | 2010 | Chinese | T2D, IGR per WHO criteria | Normal glucose regulation | rs10830963, rs1387153 | 1825 T2D, 1487 IGR | 2200 | SNaPshot |
| Hu [25] | 2010 | Chinese | T2D per WHO criteria | Normal glucose tolerant | rs10830963 | 3410 T2D | 3412 | MassARRAY |
| Kan [26] | 2010 | Chinese | T2D per WHO criteria | Normal fasting glucose | rs10830963, rs1387153 | 1912 T2D | 2041 | Taqman |
| Rees [27] | 2011 | Pakistani | T2D per WHO criteria | Normoglycaemic | rs10830963 | 1678 T2D | 1584 | KASPar |
| Dietrich [28] | 2011 | German | IGT, T2D per WHO criteria | Normal glucose tolerant | rs10830963 | 103 T2D, 73 IGT | 748 | TaqMan |
| Been [29] | 2011 | Indian | T2D per ADA criteria | Normoglycaemic | rs10830963, rs1387153 | 1169 T2D | 1001 | TaqMan |
| Ling [30] | 2011 | Chinese | T2D per WHO criteria | Non-diabetic | rs10830963 | 1118 T2D | 1161 | MassARRAY |
| Olsson [31] | 2011 | Norwegian | T2D patients | Non-diabetic | rs10830963 | 1322 T2D | 1447 | TaqMan |
| Ohshige [32] | 2011 | Japanese | T2D per WHO criteria | Non-diabetic | rs10830963, rs1387153 | 2809 T2D | 2066 | PCR-invader assay |
| Renstrom [33] | 2011 | Swedish | IFG per WHO criteria | Normal glucose regulation | rs10830963 | 964 IFG | 3087 | OpenArray |
| Song [34] | 2011 | Chinese | IFG per WHO criteria | Normal fasting glucose | rs10830963 | 288 IFG | 1742 | ARMS-PCR |
| Tabara [35] | 2011 | Japanese | T2D per ADA criteria | Non-diabetic | rs10830963, rs1387153 | 495 T2D | 399 | TaqMan |
| Liu [36] | 2012 | Chinese | T2D per ADA criteria | Normal glucose tolerant | rs1387153 | 295 T2D | 239 | RFLP |
| Walford [37] | 2012 | American | IFG per ADA criteria | Normal glucose tolerant | rs10830963 | 6251 IFG | 12480 | IBC Chip |
WHO: World Health Organization, ADA: American Diabetes Association, IFG: Impaired Fasting Glycemia, IGT: Impaired Glucose Tolerance, IGR: Impaired Glucose Regulation, OGTT: Oral Glucose Tolerance Test.
Association of MTNR1B rs10830963 polymorphism with T2D
Overall, there was evidence of an association between the increased risk of T2D and the variant in different genetic models when all the eligible studies were pooled into the meta-analysis. Using random effect model, the summary per-allele OR of the G variant for T2D was 1.05 [95% CI: 1.02–1.08, P(Z)<10−4, P(Q) = 0.01; Figure 1], with corresponding results under dominant and recessive genetic models of 1.10 [95% CI: 1.06–1.13, P(Z)<10−4, P(Q) = 0.17 ] and 1.12 [95% CI: 1.08–1.16, P(Z)<10−5, P(Q) = 0.007], respectively. In the stratified analysis by ethnicity, significantly increased risks were found among Caucasian populations [G allele: OR = 1.09, 95% CI: 1.06–1.13, P(Z)<10−5; dominant model: OR = 1.16, 95% CI: 1.07–1.25, P(Z)<10−5; recessive model: OR = 1.19, 95% CI: 1.10–1.28, P(Z)<10−5]. However, no significant association was found for East Asian and South Asian populations in all genetic models (Table 2). By considering sample size subgroups, the OR was 1.03 [95% CI: 0.97–1.09, P(Z) = 0.32, P(Q) = 0.15] in small studies compared to 1.06 [95% CI: 1.03–1.10, P(Z)<10−4, P(Q) = 0.01] in larger studies.
Figure 1. Meta-analysis of studies of the rs10830963 polymorphism of MTNR1B and T2D.
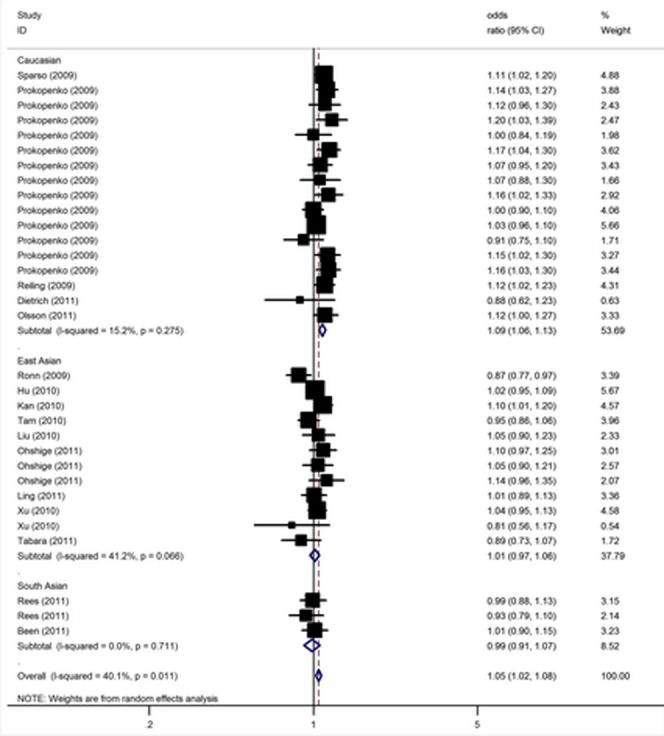
Table 2. Results of meta-analysis for MTNR1B rs10830963 polymorphism and T2D risk.
| Sub-group analysis | No. of data sets | No. of cases/controls | G Allele | Dominant Model | Recessive Model | ||||||
| OR (95%CI) | P(Z) | P(Q) | OR (95%CI) | P(Z) | P(Q) | OR (95%CI) | P(Z) | P(Q) | |||
| Overall | 32 | 41552/92618 | 1.05 (1.02–1.08) | <10−4 | 0.01 | 1.10 (1.06–1.13) | <10−4 | 0.17 | 1.12 (1.08–1.16) | <10−5 | 0.007 |
| Ethnicity | |||||||||||
| Caucasian | 17 | 24146/73443 | 1.09 (1.06–1.13) | <10−5 | 0.27 | 1.16 (1.07–1.25) | <10−5 | 0.07 | 1.19 (1.10–1.28) | <10−5 | 0.04 |
| East Asian | 12 | 14559/16590 | 1.01 (0.97–1.06) | 0.57 | 0.07 | 1.06 (0.98–1.13) | 0.11 | 0.09 | 1.08 (0.99–1.17) | 0.07 | 0.01 |
| South Asian | 3 | 2847/2585 | 0.99 (0.91–1.07) | 0.72 | 0.71 | 0.99 (0.88–1.11) | 0.81 | 0.53 | 1.07 (0.99–1.16) | 0.07 | 0.03 |
| Sample size | |||||||||||
| <1000 | 13 | 6881/18503 | 1.03 (0.97–1.09) | 0.32 | 0.15 | 1.05 (0.99–1.12) | 0.11 | 0.38 | 1.08 (0.99–1.19) | 0.09 | 0.12 |
| ≥1000 | 19 | 34671/74115 | 1.06 (1.03–1.10) | <10−4 | 0.01 | 1.11 (1.07–1.15) | <10−5 | 0.22 | 1.14 (1.09–1.19) | <10−5 | <10−4 |
In meta-regression analysis, sample size (P = 0.15), mean age of cases (P = 0.28) and controls (P = 0.16), mean BMI cases (P = 0.06) and controls (P = 0.17), sex distribution in cases (P = 0.32) and controls (P = 0.21) did not significantly explained such heterogeneity. By contrast, ethnicity (P = 0.001) was significantly correlated with the magnitude of the genetic effect, explaining 67% of the heterogeneity.
Association of MTNR1B rs10830963 polymorphism with impaired glucose regulation
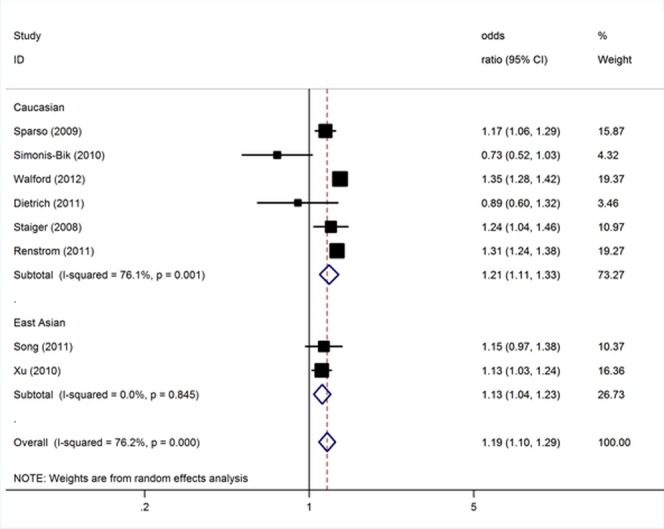
To investigate how glucose metabolism was related to MTNR1B, we analyzed individuals with impaired glucose regulation (impaired glucose tolerance and/or impaired fasting glucose). The data on genotypes of the rs10830963 polymorphism among impaired glucose regulation cases and controls were available in 8 (including 10, 810 cases and 26, 478 controls) studies [17], [18], [23], [24], [28], [33], [34], [37]. For IGR risk and the rs10830963 polymorphism of MTNR1B, our meta-analysis gave an overall per-allele OR of 1.19 (95% CI: 1.10–1.29; P<10−4) with statistically significant between-study heterogeneity (P<10−4). Significant associations were also found under dominant [OR = 1.23; 95% CI: 1.15–1.31; P(Z)<10−5; P(Q) = 0.004] and recessive [OR = 1.27; 95% CI: 1.18–1.35; P(Z)<10−5; P(Q) = 0.006] genetic model. This analysis is based on pooling of data from a number of different ethnic populations. When stratifying for ethnicity, an OR of 1.21 [95% CI: 1.11–1.33; P(Z)<10−4; P(Q)<0.001] and 1.13 [95% CI: 1.04–1.23; P(Z) = 0.003; P(Q) = 0.85] resulted for risk allele, among Caucasians and East Asians, respectively (Figure 2). By considering IGR outcome subgroups, the OR was 1.04 [95% CI: 0.87–1.25; P(Z) = 0.66; P(Q)<10−4] for IGT compared to 1.33 [95% CI: 1.17–1.52; P(Z)<10−4; P(Q) = 0.03] for IGF.
Figure 2. Meta-analysis of studies of the rs10830963 polymorphism of MTNR1B and IGR stratified by ethnicity.
Association of MTNR1B rs1387153 polymorphism with T2D
The meta-analysis resulted in a statistically non-significant association between MTNR1B rs1387153 and T2D. The overall OR for risk T allele was 1.04 [95% CI: 0.98–1.10; P(Z) = 0.20; P(Q) = 0.003; Figure 3]. Similar results were also found under dominant and recessive genetic models (Table 3). When studies were stratified for ethnicity, marginal significant associations were found among Caucasians with per-allele OR of 1.05 [95% CI: 1.00–1.11, P(Z) = 0.048; P(Q) = 0. 88]. However, no significant association was found for East Asian and South Asian populations in almost all genetic models (Table 3). Subsidiary analyses of sample size yielded a per-allele OR of 0.96 [95% CI: 0.84–1.10, P(Z) = 0.57] for small studies and for larger studies of 1.07 [95% CI: 1.00–1.14, P(Z) = 0.04].
Figure 3. Meta-analysis of studies of the rs1387153 polymorphism of MTNR1B and T2D.
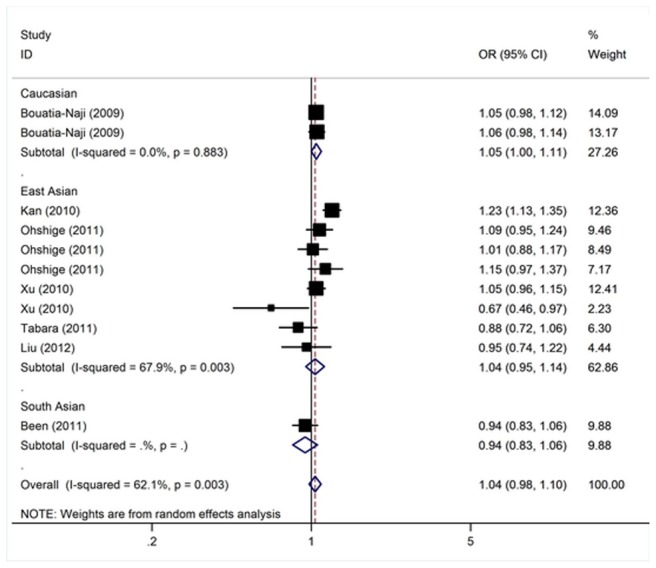
Table 3. Results of meta-analysis for MTNR1B rs1387153 polymorphism and T2D risk.
| Sub-group analysis | No. of data sets | No. of cases/controls | T Allele | Dominant Model | Recessive Model | ||||||
| OR (95%CI) | P(Z) | P(Q) | OR (95%CI) | P(Z) | P(Q) | OR (95%CI) | P(Z) | P(Q) | |||
| Overall | 11 | 14874/17703 | 1.04 (0.98–1.10) | 0.20 | 0.003 | 1.03 (0.94–1.13) | 0.55 | 0.005 | 1.11 (1.00–1.23) | 0.05 | 0.12 |
| Ethnicity | |||||||||||
| Caucasian | 2 | 6332/9132 | 1.05 (1.00–1.11) | 0.048 | 0.88 | 1.05 (0.98–1.12) | 0.14 | 0.77 | 1.12 (0.99–1.25) | 0.06 | 0.37 |
| East Asian | 8 | 7378/7598 | 1.04 (0.95–1.14) | 0.42 | 0.003 | 1.10 (1.00–1.20) | 0.05 | 0.001 | 1.18 (1.06–1.31) | 0.004 | 0.11 |
| South Asian | 1 | 1164/973 | 0.94 (0.83–1.06) | 0.32 | NA | 0.91 (0.77–1.09) | 0.31 | NA | 0.93 (0.72–1.20) | 0.57 | NA |
| Sample size | |||||||||||
| <1000 | 5 | 2045/2690 | 0.96 (0.84–1.10) | 0.57 | 0.06 | 0.93 (0.82–1.06) | 0.28 | 0.16 | 1.07 (0.91–1.26) | 0.44 | 0.19 |
| ≥1000 | 6 | 12829/15013 | 1.07 (1.00–1.14) | 0.04 | 0.01 | 1.08 [1.03, 1.14] | 0.004 | 0.01 | 1.14 (1.05–1.24) | 0.002 | 0.12 |
NA: Not Available.
In meta-regression analysis, neither sample size (P = 0.37), ethnicity (P = 0.46), mean BMI of cases (P = 0.17) and controls (P = 0.29), mean age of cases (P = 0.09) and controls (P = 0.67), nor sex distribution in cases (P = 0.39) and controls (P = 0.11) were significantly correlated with the magnitude of the genetic effect.
Sensitivity analyses and publication bias
A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data-set to the pooled ORs, and the corresponding pooled ORs were not qualitatively altered (Figure S2 and S3). Begg's funnel plot and Egger's test were performed to access the publication bias of the literatures. The shape of the funnel plots was symmetrical for these polymorphisms (Figure S4 and S5). The statistical results still did not show publication bias in these studies for rs10830963 (Egger test, P = 0.38; Figure S6) and rs1387153 (Egger test, P = 0.12; Figure S7).
Discussion
Large sample and unbiased epidemiological studies of predisposition genes polymorphisms could provide insight into the in vivo relationship between candidate genes and diseases. GWAS and meta-analysis have shown that common variation in the MTNR1B (rs10830963, rs1387153) locus increases the level of FPG. However, the relationship between these common variations and T2D susceptibility has not been built up yet. This is the first comprehensive meta-analysis examined the MTNR1B polymorphisms (rs10830963, rs1387153) and the relationship to T2D risk. Its strength was based on the accumulation of published data giving greater information to detect significant differences. In total, the present meta-analysis combined 23 studies including 48, 278 T2D cases, 10,810 IGR cases, and 119,960 controls.
Our results demonstrated that the rs10830963 polymorphism of MTNR1B is a risk factor for developing type 2 diabetes. In the stratified analysis by ethnicity, significant associations were found in Caucasians for the polymorphism in all genetic models. However, no significant associations were detected among East Asian and South Asian populations for rs10830963 and rs1387153 polymorphisms. There are several possible reasons for such differences. Firstly, the frequencies of the risk-association alleles in MTNR1B vary between different races. For example, the G allele distributions of the rs10830963 polymorphism varies between East Asian, South Asian and Caucasian populations, with a prevalence of ∼42%, ∼39%, and ∼25%, respectively. Therefore, additional studies are warranted to further validate ethnic difference in the effect of these polymorphisms on TD risk. Secondly, such different results could also be explained by study design or sample size. Besides, other confounding factors, such as age, sex, life style should also be considered. In the stratified analysis according to sample size, significantly associations were found only found for larger studies. Thus, absence of association with type 2 diabetes in small study could be due to insufficient power. Thus, for future association studies much larger sample size will be required.
Melatonin is mainly produced by the pineal gland but is also released from the gastrointestinal tract [38]. As a highly lipophilic circulating hormone, melatonin easily reaches and penetrates all cells and, in addition to controlling circadian rhythm, it has the ability to neutralise reactive oxygen and nitrogen species and activate the immune system [39]. Several studies have shown a link between disturbances of circadian rhythm and metabolic diseases, including diabetes [40], [41], as well as a clear relationship between insulin and melatonin [42]. Two distinct G protein-coupled receptors, MTNR1A and MTNR1B, mediate the effects of melatonin. These two receptors have been found to be produced in human pancreatic islets [42], [43], and the levels of both are upregulated in type 2 diabetic patients [44]. Furthermore, Lyssenko et al. [3] confirmed the presence of MTNR1B in human pancreatic islets and showed increased MTNR1B mRNA expression in carriers of the rs10830963 risk genotype, reporting a negative correlation between MTNR1B mRNA levels and insulin secretion. The recent finding that MTNR1B is expressed in the β-cells implies that the gene variant might affect pancreatic glucose sensing and/or insulin release and thereby glucose tolerance [4]. However, rs10830963 is located in the intronic region of MTNR1B, while rs1387153 is located in the 5′ region. These variants might be involved in regulation of promoter activity, but additional analyses of these SNPs are required to provide clear evidence on their functional consequences. Candidate gene based studies showed that focusing on functionally significant alleles can increase statistical signal and, hence, the power to detect association between presence of rare variants and complex traits. Recently, Bonnefond et al. reported that rare MTNR1B variants impairing melatonin receptor 1B function contribute to T2D [45], implying that the previously observed increased MTNR1B expression may not be causal.
Impaired glucose regulation (IGR) includes impaired fasting glucose (IFG) and/or impaired glucose tolerance (IGT). IGR is also known as intermediate hyperglycemia or pre-diabetes and characterized by high blood glucose concentrations, insulin resistance and impaired insulin secretion. Previous studies have shown that 5∼10% IGT subjects developed diabetes each year, although, some of them could revert spontaneously to normal glucose tolerance [46], [47]. However, few studies were concerned about the association of those GWAS variations with IGR [48]. IFG and/or IGT were predisposed to diabetes; however, whether the IGR and T2DM shared the same spectrum of genetic variations is not well characterized. Here in our study, we found that rs10830963 of MTNR1B that are associated with T2D was also conferred the risk of IGR. Our study suggested that IGR might have similar background of susceptible genetic variations. In addition, our results indicated that significantly increased risk of MTNR1B rs10830963 polymorphism was found for IGF but not for IGT when stratified by IGR outcome. However, because the IGR included IFG and IGT which may have different genetic etiology [5], [49], more prospectively-designed association studies with large sample size and homogeneous patients are needed in the near future.
In interpreting the results, some limitations of this meta-analysis should be addressed. Firstly, the subgroup meta-analyses on Asian populations are based on a small number of studies with such information available. Nevertheless, the total number of subjects included in this part of the analysis comprises the largest sample size so far. As studies among the Non-Caucasians are currently limited, further studies including a wider spectrum of subjects should be carried to investigate the role of these variants in different populations. Secondly, our results were based on unadjusted estimates, while a more precise analysis should be conducted if all individual raw data were available, which would allow for the adjustment by other co-variants including age, drinking status, obesity, cigarette consumption, and other lifestyle. Thirdly, heterogeneity is a potential problem when interpreting all the results of meta-analysis. Although we minimized the likelihood by performing a careful search for published studies, using the explicit criteria for study inclusion, the significant between-study heterogeneity still existed in most of comparison. Besides, subgroup analysis and meta-regression were also used to identify the source of heterogeneity. The presence of heterogeneity can result from differences in the age distribution, obesity status of subjects, selection of controls, dietary habits, prevalence lifestyle factors and so on. Last but not least, only published studies were included in this meta-analysis. Therefore, publication bias may have occurred, even though the use of a statistical test did not show it.
To conclude, this meta-analysis showed that the MTNR1B rs10830963 polymorphism was significantly associated with increased risk of T2D, particularly in the Caucasian population. In addition, our meta-analysis results suggest that MTNR1B rs10830963 is risk factor for the development of impaired glucose regulation. Moreover, gene–gene and gene–environment interactions should be considered in future studies.
Supporting Information
(DOC)
Study selection process.
(TIF)
Result of sensitivity analyses for MTNR1B rs10830963.
(TIF)
Result of sensitivity analyses for MTNR1B rs1387153.
(TIF)
Begg's funnel plot of MTNR1B rs10830963 polymorphism and T2D risk.
(TIF)
Begg's funnel plot of MTNR1B rs1387153 polymorphism and T2D risk.
(TIF)
Test publication bias of studies of the rs10830963 polymorphism of MTNR1B and T2D using Egger test.
(TIF)
Test publication bias of studies of the rs1387153 polymorphism of MTNR1B and T2D using Egger test.
(TIF)
Funding Statement
This work was supported by China Postdoctoral Science Foundation Funded Project (20100480542, 201104227), Nature Science Foundation of Shanghai (12ZR1436000) and Youth Innovation Promotion Association, Chinese Academy of Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Watanabe RM, Valle T, Hauser ER, Ghosh S, Eriksson J, et al. (1999) Familiality of quantitative metabolic traits in Finnish families with non-insulin-dependent diabetes mellitus. Finland-United States Investigation of NIDDM Genetics (FUSION) Study investigators. Hum Hered 49: 159–168. [DOI] [PubMed] [Google Scholar]
- 2. Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, et al. (2009) Variants in MTNR1B influence fasting glucose levels. Nat Genet 41: 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, et al. (2009) Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 41: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, et al. (2009) A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 41: 89–94. [DOI] [PubMed] [Google Scholar]
- 5. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, et al. (2008) A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science 320: 1085–1088. [DOI] [PubMed] [Google Scholar]
- 7. Peschke E, Bach AG, Muhlbauer E (2006) Parallel signaling pathways of melatonin in the pancreatic beta-cell. J Pineal Res 40: 184–191. [DOI] [PubMed] [Google Scholar]
- 8. Mulder H, Nagorny CL, Lyssenko V, Groop L (2009) Melatonin receptors in pancreatic islets: good morning to a novel type 2 diabetes gene. Diabetologia 52: 1240–1249. [DOI] [PubMed] [Google Scholar]
- 9. Muhlbauer E, Gross E, Labucay K, Wolgast S, Peschke E (2009) Loss of melatonin signalling and its impact on circadian rhythms in mouse organs regulating blood glucose. Eur J Pharmacol 606: 61–71. [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, Martin CC, Oeser JK, Sarkar S, McGuinness OP, et al. (2007) Deletion of the gene encoding the islet-specific glucose-6-phosphatase catalytic subunit-related protein autoantigen results in a mild metabolic phenotype. Diabetologia 50: 774–778. [DOI] [PubMed] [Google Scholar]
- 11. Cochran WG (1954) The combination of estimates from different experiments. Biometrics 10: 101–129. [Google Scholar]
- 12. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 13. Thompson SG, Sharp SJ (1999) Explaining heterogeneity in meta-analysis: a comparison of methods. Stat Med 18: 2693–2708. [DOI] [PubMed] [Google Scholar]
- 14. Woolf B (1955) On estimating the relation between blood group and disease. Ann Hum Genet 19: 251–253. [DOI] [PubMed] [Google Scholar]
- 15. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101. [PubMed] [Google Scholar]
- 17. Staiger H, Machicao F, Schäfer SA, Kirchhoff K, Kantartzis K, et al. (2008) Polymorphisms within the novel type 2 diabetes risk locus MTNR1B determine beta-cell function. PLoS One 3: e3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sparsø T, Bonnefond A, Andersson E, Bouatia-Naji N, Holmkvist J, et al. (2009) G-allele of intronic rs10830963 in MTNR1B confers increased risk of impaired fasting glycemia and type 2 diabetes through an impaired glucose-stimulated insulin release: studies involving 19,605 Europeans. Diabetes 58: 1450–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rönn T, Wen J, Yang Z, Lu B, Du Y, et al. (2009) A common variant in MTNR1B, encoding melatonin receptor 1B, is associated with type 2 diabetes and fasting plasma glucose in Han Chinese individuals. Diabetologia 52: 830–833. [DOI] [PubMed] [Google Scholar]
- 20. Reiling E, van 't Riet E, Groenewoud MJ, Welschen LM, van Hove EC, et al. (2009) Combined effects of single-nucleotide polymorphisms in GCK, GCKR, G6PC2 and MTNR1B on fasting plasma glucose and type 2 diabetes risk. Diabetologia 52: 1866–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tam CH, Ho JS, Wang Y, Lee HM, Lam VK, et al. (2010) Common polymorphisms in MTNR1B, G6PC2 and GCK are associated with increased fasting plasma glucose and impaired beta-cell function in Chinese subjects. PLoS One 5: e11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu C, Wu Y, Li H, Qi Q, Langenberg C, et al. (2010) MTNR1B rs10830963 is associated with fasting plasma glucose, HbA1C and impaired beta-cell function in Chinese Hans from Shanghai. BMC Med Genet 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simonis-Bik AM, Nijpels G, van Haeften TW, Houwing-Duistermaat JJ, Boomsma DI, et al. (2010) Gene variants in the novel type 2 diabetes loci CDC123/CAMK1D, THADA, ADAMTS9, BCL11A, and MTNR1B affect different aspects of pancreatic beta-cell function. Diabetes 59: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu M, Bi Y, Xu Y, Yu B, Huang Y, et al. (2010) Combined effects of 19 common variations on type 2 diabetes in Chinese: results from two community-based studies. PLoS One 5: e14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu C, Zhang R, Wang C, Yu W, Lu J, et al. (2010) Effects of GCK, GCKR, G6PC2 and MTNR1B variants on glucose metabolism and insulin secretion. PLoS One 5: e11761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kan MY, Zhou DZ, Zhang D, Zhang Z, Chen Z, et al. (2010) Two susceptible diabetogenic variants near/in MTNR1B are associated with fasting plasma glucose in a Han Chinese cohort. Diabet Med 27: 598–602. [DOI] [PubMed] [Google Scholar]
- 27. Rees SD, Hydrie MZ, O'Hare JP, Kumar S, Shera AS, et al. (2011) Effects of 16 genetic variants on fasting glucose and type 2 diabetes in South Asians: ADCY5 and GLIS3 variants may predispose to type 2 diabetes. PLoS One 6: e24710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dietrich K, Birkmeier S, Schleinitz D, Breitfeld J, Enigk B, et al. (2011) Association and evolutionary studies of the melatonin receptor 1B gene (MTNR1B) in the self-contained population of Sorbs from Germany. Diabet Med 28: 1373–1380. [DOI] [PubMed] [Google Scholar]
- 29. Been LF, Hatfield JL, Shankar A, Aston CE, Ralhan S, et al. (2011) A low frequency variant within the GWAS locus of MTNR1B affects fasting glucose concentrations: Genetic risk is modulated by obesity. Nutr Metab Cardiovasc Dis doi:10.1016/j.numecd.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ling Y, Li X, Gu Q, Chen H, Lu D, Gao X (2011) A common polymorphism rs3781637 in MTNR1B is associated with type 2 diabetes and lipids levels in Han Chinese individuals. Cardiovasc Diabetol 10: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Olsson L, Pettersen E, Ahlbom A, Carlsson S, Midthjell K, et al. (2011) No effect by the common gene variant rs10830963 of the melatonin receptor 1B on the association between sleep disturbances and type 2 diabetes: results from the Nord-Trondelag Health Study. Diabetologia 54: 1375–1378. [DOI] [PubMed] [Google Scholar]
- 32. Ohshige T, Iwata M, Omori S, Tanaka Y, Hirose H, et al. (2011) Association of new loci identified in European genome-wide association studies with susceptibility to type 2 diabetes in the Japanese. PLoS One 6: e26911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Renström F, Shungin D, Johansson I (2011) MAGIC Investigators (2011) Florez JC, et al. (2011) Genetic predisposition to long-term nondiabetic deteriorations in glucose homeostasis: Ten-year follow-up of the GLACIER study. Diabetes 60: 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song JY, Wang HJ, Ma J, Xu ZY, Hinney A, et al. (2011) Association of the rs10830963 polymorphism in MTNR1B with fasting glucose levels in Chinese children and adolescents. Obes Facts 4: 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tabara Y, Osawa H, Kawamoto R, Onuma H, Shimizu I, et al. (2011) Genotype risk score of common susceptible variants for prediction of type 2 diabetes mellitus in Japanese: the Shimanami Health Promoting Program (J-SHIPP study). Development of type 2 diabetes mellitus and genotype risk score. Metabolism 60: 1634–1640. [DOI] [PubMed] [Google Scholar]
- 36. Liu Y, Zhou L, Xie XM, Yang Z (2012) The correlation of MTNR1B gene polymorphism with type 2 diabetes mellitus in Ningxia Han population. Zhongguo Lao Nian Xue Zazhi 6: 111–1121. [Google Scholar]
- 37. Walford GA, Green T, Neale B, Isakova T, Rotter JI, et al. (2012) Common genetic variants differentially influence the transition from clinically defined states of fasting glucose metabolism. Diabetologia 55: 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Konturek SJ, Konturek PC, Brzozowski T, Bubenik GA (2007) Role of melatonin in upper gastrointestinal tract. J Physiol Pharmacol 58 Suppl 6: 23–52. [PubMed] [Google Scholar]
- 39. Jaworek J, Nawrot-Porabka K, Leja-Szpak A, Bonior J, Szklarczyk J, et al. (2007) Melatonin as modulator of pancreatic enzyme secretion and pancreatoprotector. J Physiol Pharmacol 58 Suppl 6: 65–80. [PubMed] [Google Scholar]
- 40. Knutson KL, Spiegel K, Penev P, Van Cauter E (2007) The metabolic consequences of sleep deprivation. Sleep Med Rev 11: 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laposky AD, Bass J, Kohsaka A, Turek FW (2008) Sleep and circadian rhythms: key components in the regulation of energy metabolism. FEBS Lett 582: 142–151. [DOI] [PubMed] [Google Scholar]
- 42. Peschke E (2008) Melatonin, endocrine pancreas and diabetes. J Pineal Res 44: 26–40. [DOI] [PubMed] [Google Scholar]
- 43. Ramracheya RD, Muller DS, Squires PE, Brereton H, Sugden D, et al. (2008) Function and expression of melatonin receptors on human pancreatic islets. J Pineal Res 44: 273–279. [DOI] [PubMed] [Google Scholar]
- 44. Peschke E, Stumpf I, Bazwinsky I, Litvak L, Dralle H, et al. (2007) Melatonin and type 2 diabetes — a possible link? J Pineal Res 42: 350–358. [DOI] [PubMed] [Google Scholar]
- 45. Bonnefond A, Clément N, Fawcett K, Yengo L, Vaillant E, et al. (2012) Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet 44: 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gerstein HC, Santaguida P, Raina P, Morrison KM, Balion C, et al. (2007) Annual incidence and relative risk of diabetes in people with various categories of dysglycemia: a systematic overview and meta-analysis of prospective studies. Diabetes Res Clin Pract 78: 305–312. [DOI] [PubMed] [Google Scholar]
- 47. Saad MF, Knowler WC, Pettitt DJ, Nelson RG, Mott DM, et al. (1988) The natural history of impaired glucose tolerance in the Pima Indians. N Engl J Med 319: 1500–1506. [DOI] [PubMed] [Google Scholar]
- 48. Wang J, Kuusisto J, Vänttinen M, Kuulasmaa T, Lindström J, et al. (2007) Variants of transcription factor 7-like 2 (TCF7L2) gene predict conversion to type 2 diabetes in the Finnish Diabetes Prevention Study and are associated with impaired glucose regulation and impaired insulin secretion. Diabetologia 50: 1192–1200. [DOI] [PubMed] [Google Scholar]
- 49. Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, et al. (2010) Genetic variation in GIPR influences glucose and insulin responses to an oral glucose challenge. Nat Genet 42: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Study selection process.
(TIF)
Result of sensitivity analyses for MTNR1B rs10830963.
(TIF)
Result of sensitivity analyses for MTNR1B rs1387153.
(TIF)
Begg's funnel plot of MTNR1B rs10830963 polymorphism and T2D risk.
(TIF)
Begg's funnel plot of MTNR1B rs1387153 polymorphism and T2D risk.
(TIF)
Test publication bias of studies of the rs10830963 polymorphism of MTNR1B and T2D using Egger test.
(TIF)
Test publication bias of studies of the rs1387153 polymorphism of MTNR1B and T2D using Egger test.
(TIF)


