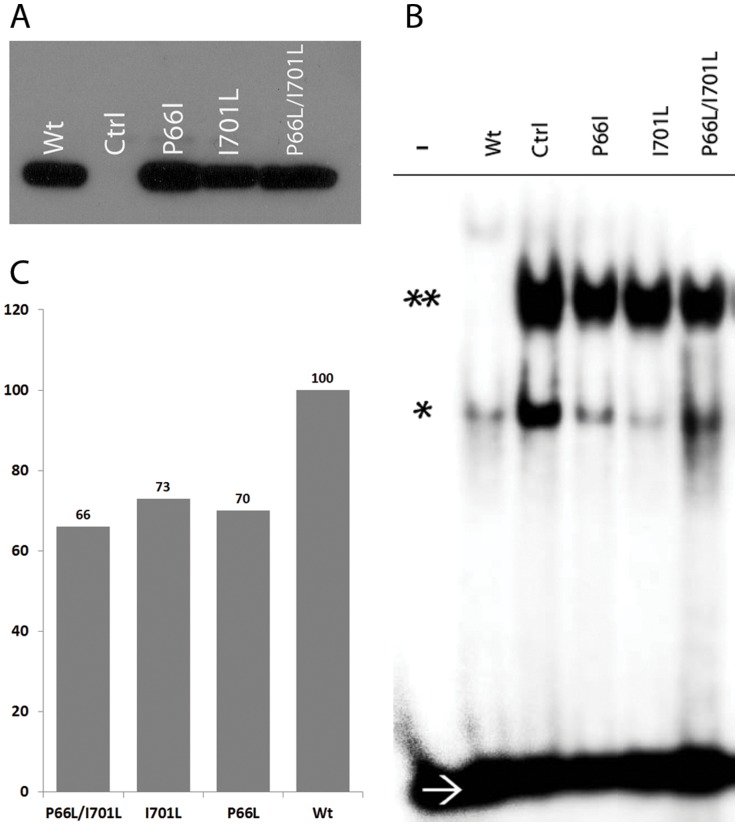
Figure 5. DNA binding affinity of the mutated NFATC1 proteins.
A- NFATC1 extracts from HEK 293 cells transfected with Wt NFATC1 and Mutants (P66L – I701L – P66L/I701L) were resolved on an SDS-PAGE prior to gel shift assays. Western blots showed equal amounts of expressed proteins as depicted by the anti-Flag antibody. (Ctrl refers to nuclear extracts from mock-transfected cells). B- EMSA was performed using equal amounts of the overexpressed NFATC1 proteins from HEK 293 cells transfected with Wt NFATC1 and NFATC1 mutants (P66L, I701L, P66L/I701L) and NFAT-consensus binding site as a probe. – ve sign indicates absence of nuclear extracts/* indicates NFATC1 monomer/** indicates NFATC1 Dimer/→ refers to the 32P labeled free DNA probe. C- Quantification of the NFATC1 dimers in the EMSA using the TotalLab2010 software from Amersham shows a 30% decrease in DNA binding affinity of the single and double mutant as compared to the wild type NFATC1 protein.