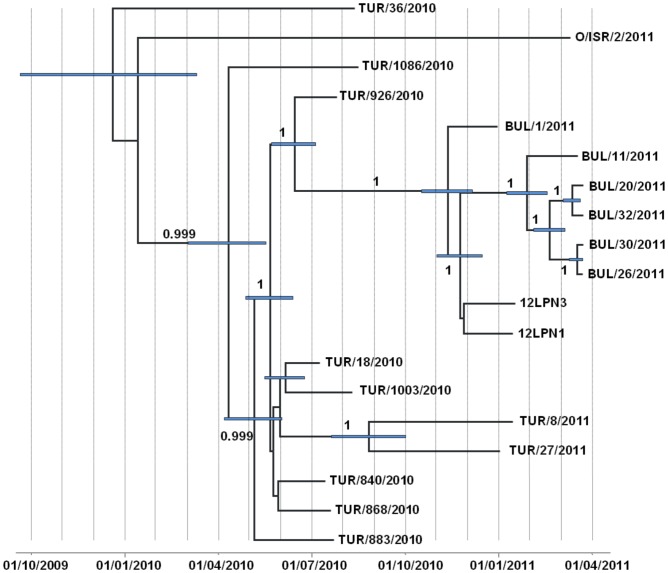
Figure 4. Bayesian maximum-clade-credibility time-scaled phylogenetic tree (BEAST) generated using 19 sequenced FMDV full genomes.
The analysis was undertaken using a GTR substitution model, relaxed clock, constant population size, sampling 30.000 trees from 30 million generations. Uncertainty for the date of each node (95% highest posterior density – HPD - intervals) is displayed in bars. Only node labels with posterior over 0.8 are indicated. Overall, a rate of nucleotide substitution of 9.05×10−3 (95% HPD: 6.99–11.11×10−3) per site per year was estimated.