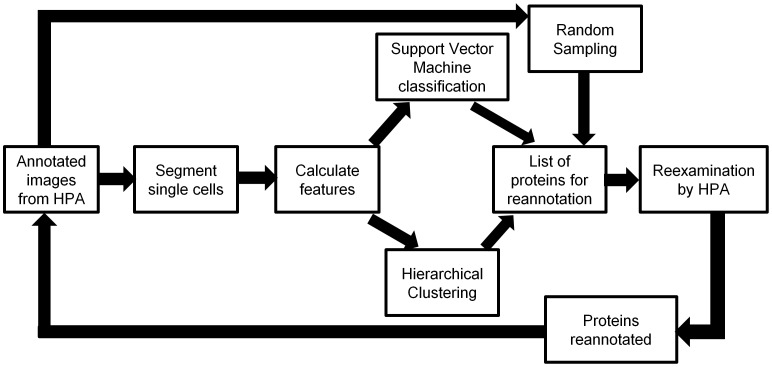
Figure 1. An overview of the framework introduced in this paper.
We first collect immunofluorescence images from confocal microscopy that are annotated visually by HPA. These image fields are then segmented into single cells and various features (SLFs) are calculated. We then do two parallel analyses on the features. One is a supervised classification using a Support Vector Machine, and the other is an unsupervised hierarchical clustering. This results in two lists of proteins which have high probabilities of needing reannotation. Combined with another list of proteins sampled randomly, a final list which contains only the protein IDs are reexamined by the HPA annotation group. The annotations of some proteins may change, and these modified annotations can be incorporated in another cycle of analysis.