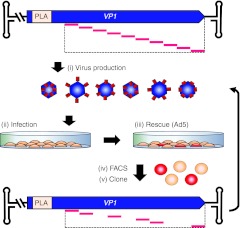
Figure 2.
Selection for fluorescent VP1-mCherry hybrids. (i) The pAAV2_RaPID library was used to produce a pool of AAV2 hybrids with mCherry inserted in the VP3 region of VP1 only, which contains the PLA2 domain required for infectivity; wt VP2/3 genes were supplemented in trans. (ii) A monolayer of HEK-293T cells were infected with the virus pool. (iii) Successfully infectious variants were then rescued by superinfection with Ad5. (iv) Cells were harvested and sorted by FACS to isolate those expressing fluorescent VP1-mCherry proteins. (v) DNA was extracted from the sorted cells, amplified by PCR, and subcloned back into the VP1 expression vector, generating an enriched library gene pool. The selection procedure was iterated twice more using the enriched gene pools for a total of three rounds of selection. FACS, fluorescence-activated cell sorting.