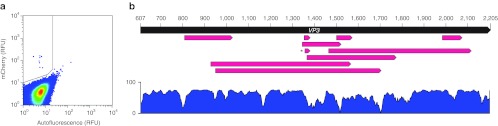
Figure 3.
Screening of fluorescent VP1-mCherry hybrids isolated after one round of selection. (a) FACS data from round 1 selection. Y-axis indicates the mCherry signal in relative fluorescence units (RFU). X-axis represents cell autofluorescence in the green spectrum. Gate in upper left quadrant defines the region where cells were collected during selection. (b) Clones recovered after the first round of selection are illustrated as in Figure 2. Each clone is represented by a red arrow that indicates the region of VP3 deleted at the site of mCherry insertion. Clone r1c3 is marked with an asterisk. A similarity plot is shown below, indicating the regions of VP3 that are conserved across AAV serotypes 1-12, generated from an alignment in Vector NTI. Bar at left indicates percent similarity. Note r1c3 lies in a wide valley of the similarity plot located at the GH2/3 loop. AAV, adeno-associated virus; FACS, fluorescence-activated cell sorting.