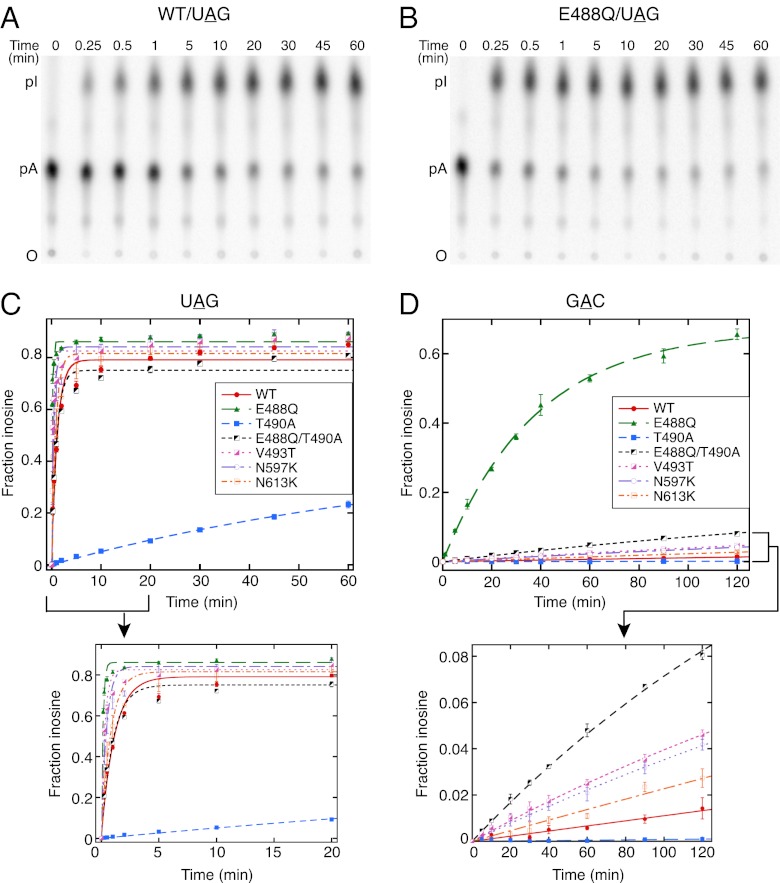
Fig. 3.
The kdeam values for some mutant enzymes are similar to that of WT hADAR2, but others differ. (A and B) PhosphorImages showing representative TLC plates used in the kdeam assay with 250 nM WT hADAR2 (A) or E488Q mutant (B) and 0.5 nM UAG hairpin with the target adenosine labeled at its 5′ phosphate. Time points are indicated at the top of the TLC plate, positions of origin (O), 5′ AMP (pA), and 5′ IMP (pI) are indicated on the left. Control experiments using less protein or twice the amount of RNA confirmed single-turnover conditions (Fig. S2) and also established that WT and mutant hADAR2 were stable for the duration of the experiment (Fig. S3). (C and D) Plots showing the fraction of inosine produced as a function of time for WT hADAR2 and mutants with UAG (C) and GAC (D) hairpins. Data points were fitted to the equation, Ft = Fend (1 − e−kt), where Ft is the fraction of inosine at time t, Fend is the fitted fraction of inosine at end point, and k is the fitted rate constant. Error bars indicate SD; n ≥ 3. Insets expand the x-axis for reactions with the UAG hairpin and the y-axis for reactions with the GAC hairpin. Although the overall fit to this equation was good, late time points showed a continued increase in inosine for the UAG hairpin. This continued increase could indicate a double-exponential rate, but the kdeam values obtained on excluding the late time points by fitting the data points up to 30 min were similar to that obtained from 60-min time points. The small increase at later time points possibly was caused by slow editing of contaminating 32P 5′-end–labeled 54-nt RNA used as starting material for preparing the 60-nt UAG hairpin by splint ligation.