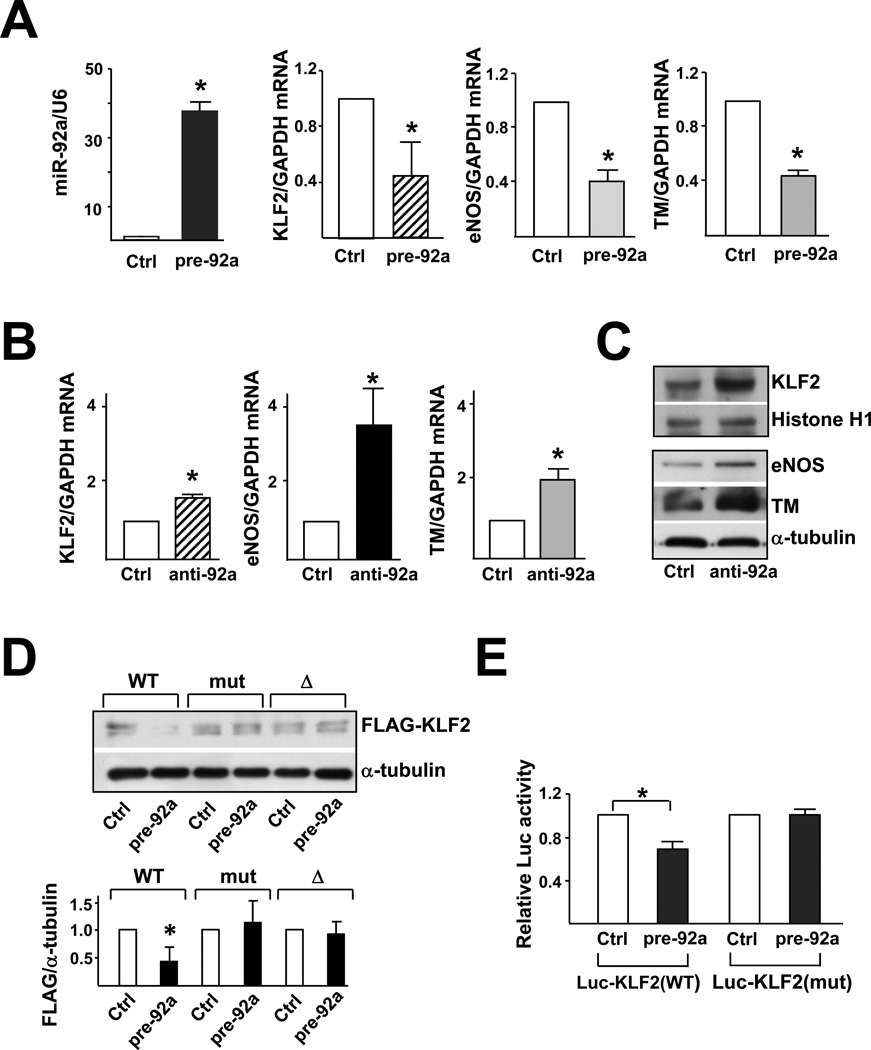
Figure 2. miR-92a targets KLF2 mRNA and decreases KLF2 translation.
HUVECs were transfected with 20 nM miR-92a precursor (pre-92a) or control RNA. At 48 hr, the levels of miR-92a relative to U6 RNA and KLF2, eNOS, and TM mRNA as ratios to GAPDH were assessed by qRT-PCR. In (B) and (C), HUVECs were transfected with 20 nM miR-92a inhibitor (anti-92a) or control RNA. The mRNA levels of KLF2, eNOS, and TM as ratios to GAPDH were assessed by qRT-PCR. T h e protein levels of KLF2, Histone H1 and eNOS were determined by Western blotting. (D) HEK293 cells were transfected with the wild-type FLAGKLF2 (WT), FLAG-KLF2 (mut) (miR-92a binding site mutation), or Flag-KLF2 (Δ) (miR-92a 24 binding site deletion) together with 20 nM pre-92a or control RNA for 48 hr. The cells were then lysed, and the level of exogenously expressed FLAG-KLF2 fusion proteins was detected by Western blot analysis with anti-FLAG. Shown in the bottom panel is the densitometry analysis of the protein amount normalized to that in the control RNA-transfected cells. (E) HEK293 cells were transfected with the wild-type Luc-KLF2-3’UTR(WT) or Luc-KLF2-3’UTR(mut) together with 20 nM pre-92a or control RNA and CMV-β-gal. The luciferase activity was normalized to that of β-gal. The data represent mean±SD from 3 independent experiments. * p<0.05 between cells transfected with pre-92a and control RNA.