Figure 2.
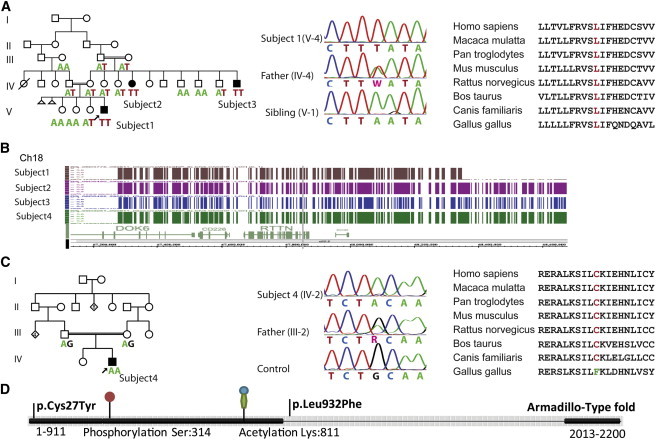
Identification of the RTTN Mutations in Families with PMG
(A) Left, pedigree of family 1. Affected subjects have black symbols, and below, the RTTN genotype is indicated, which illustrates cosegregation of the c.2796A>T change with the phenotype. Middle, electropherogram showing the homozygous mutation (subject V-4), the heterozygous father (IV-4), and the normal sequence (healthy sibling V-1). PCR products were purified with ExoSAP-IT (USB), and direct sequencing of both strands was performed with BigDye Terminator chemistry v.3.1 on an ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems). Sequences were aligned and compared with consensus sequences obtained from the human genome databases (SeqScape v.2.5 software, Applied Biosystems). For annotation of DNA and protein changes, the mutation nomenclature guidelines from the Human Genome Variation Society were followed. Right, conservation of leucine 932 among species.
(B) Visualization with the Genotyping Console browser of Affymetrix SNP 6.0 Array data, showing the overlapping areas of homozygosity among individuals from family 1 (upper three) and family 2 (lowest lane).
(C) Left, pedigree of family 2. The affected subject has a black symbol, and below, the RTTN genotype at position c.80 is indicated. Middle, electropherogram showing the homozygous mutation (subject IV-2), the heterozygous father (III-2), and the normal sequence (control). Right, conservation of cysteine 27 among species.
(D) Schematic representation of Rotatin, including the position of the amino acid change, Armadillo-like domains, and posttranslational modification sites.
The c.2796A>T change was not present in 100 ethnically matched, healthy Turkish individuals and 165 individuals of European descent. The c.80G>A change was absent in 98 Cape Verdean ethnically matched individuals and 166 healthy individuals of European descent. Both the c.2796A>T and the c.80G>A changes were not annotated as polymorphic in dbSNP130, nor were they present in 679 control individuals of the 1000 Genomes database and the Exome Variant Sever (University of Washington), indicating an extremely low allele frequency in the healthy population. Algorithms PolyPhen-2, SNAP, and Mutation Taster predicted the changes as probably being deleterious (see Web Resources).
