Figure 3.
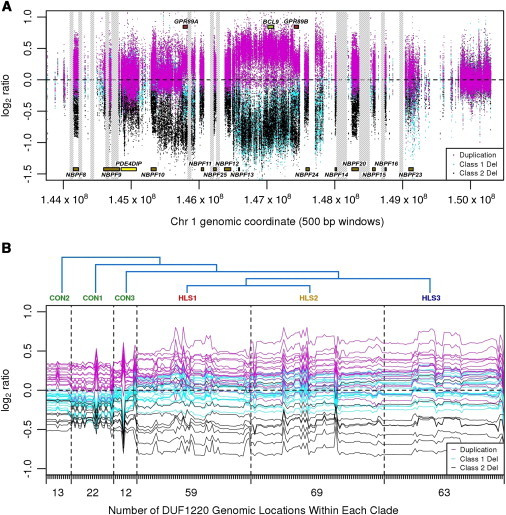
aCGH Data for Three Types of Disease Samples
Class I duplications are shown in magenta, class I deletions are shown in teal, and class II deletions are shown in black.
(A) Array data for the entire 1q21.1-1q21.2 region. Sequence gaps are displayed as vertical gray bars. Because of space limitations, only the NBPF gene family, BCL9, and GPR89 are labeled.
(B) DUF1220 clade array profile. Log2 ratios corresponding to DUF1220 signals are shown for individual samples from the three classes of the disease population. aCGH data for each individual are plotted as a continuous horizontal line so that results from individual samples can be distinguished from one another. For each sample tested, data points are first grouped according to clade and then within each clade, they are grouped in the order in which they occur across the 1q21 region. The numbers of DUF1220 domains belonging to each clade are represented as tick marks along the x axis. The number displayed below represents the number of DUF1220 domains found within each clade. The marks represent 238 clade-specific DUF1220 domains located in the 1q21 region and present on the custom array. Phylogeny of the six clades (CON1–CON3 and HLS1–HLS3) is displayed at the top.
