Figure 2.
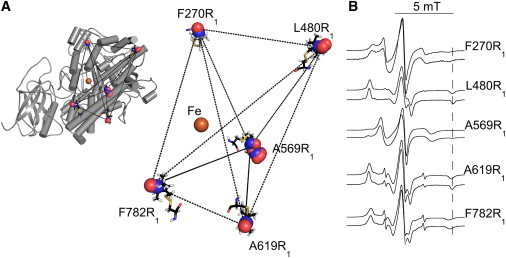
Experimental strategy. (A) Placement of the spin-labeled R1 side chain, replacing natural side chains, in the structure of soybean seed lipoxygenase-1 (SBL1) (PDB:1YGE). The coordinates of spin-labeled residues F270R1, L480R1, A519R1, A619R1, and F782R1 were generated with the software PRONOX. The allowed solutions for nitroxide oxygen (red) and nitrogen (blue) are illustrated (spheres), and one full nitroxide at each site is rendered (sticks). The catalytic iron ion is indicated (orange). Representative distances (lines) between spin labels to be measured by PDS. (Left) Placement of the spin-label sites in the overall SBL1 structure. Helices that contain <5 amino acids are simplified (loops), modifying PDB:1YGE from 46 to 26 helices. (Right) The calculated spin locations are enlarged. (B) Solution EPR spectra of SBL1 spin labeled at single sites. The maximum hyperfine separations, qualitative measures of R1 side-chain vary (verticle line drawn on the right side of the figure). Samples were at pH 8.4 (0.1 M TRICINE-HCl, 22°C), without (upper) and with (lower) 30% sucrose (w/v).
