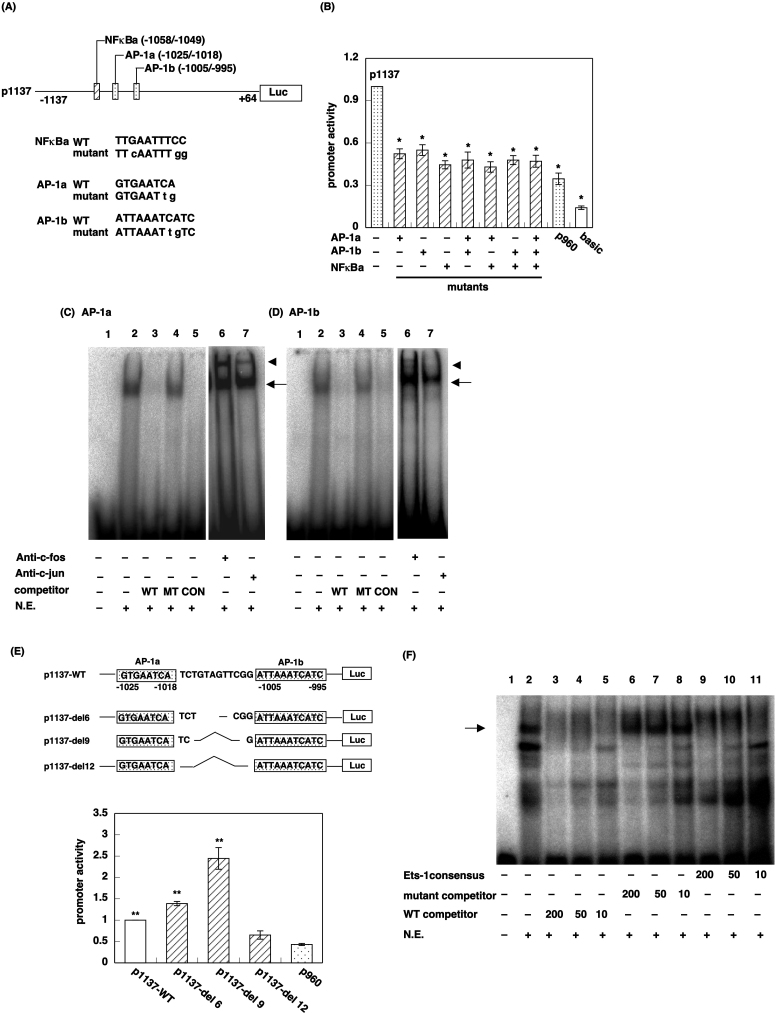
Figure 2. Identification of the transcription factor bound to the minimum sequence responsible for promoter activity in region A.
(A) Upper panel: Schematic diagram of region A, which contains one putative NF-κB binding site (NF-κBa) and two putative Ap-1 binding sites (AP-1a and AP-1b). Mutations are indicated by lowercase letters. Lower panel; Nucleotide sequences of WT and MT oligonucleotides for the three putative transcription factor binding sites. (B) Luciferase reporter assay. Data are presented as mean ± S.E.M. (n = 8). *, p < 0.01 vs p1137. (C and D) EMSA of AP-1a (C) and AP-1b (D). The nuclear extract (3 μg protein) was incubated with 32P-labeled probe bearing AP-1a (C) and Ap-1b (D), putative AP-1 binding sites. For competition studies, 200-fold molar excess of unlabeled WT (lane 3), MT (lane 4), or Ap-1 CON sequence (lane 5) oligonucleotides were used. For the supershift assay, the nuclear extract was incubated with 32P-labeled probe with or without anti-c-Fos (lane 6) or anti-c-Jun (lane 7) antibody. The positions of the sequence-specific DNA–protein complex and the shifted band are shown as an arrow and arrowhead, respectively. (E) The effect of deletion between the two AP-1 binding sites on H1R promoter activity. Upper panel: Schematic diagram of serial deletion MTs. Lower panel: Luciferase assay. Data are presented as mean ± S.E.M. (n = 6). *, p < 0.01 vs p960. (F) Ets-1 but not NF-κB binds to the NF-κBa site. The nuclear extract (3 μg protein) was incubated with 32P-labeled probe bearing NF-κBa binding site with or without 10–200-fold molar excess of unlabeled WT (lanes 3–5), MT (lanes 6–8), or Ets-1 CON sequence (lanes 9–11) oligonucleotides. The position of the sequence-specific DNA–protein complex is shown as an arrow.