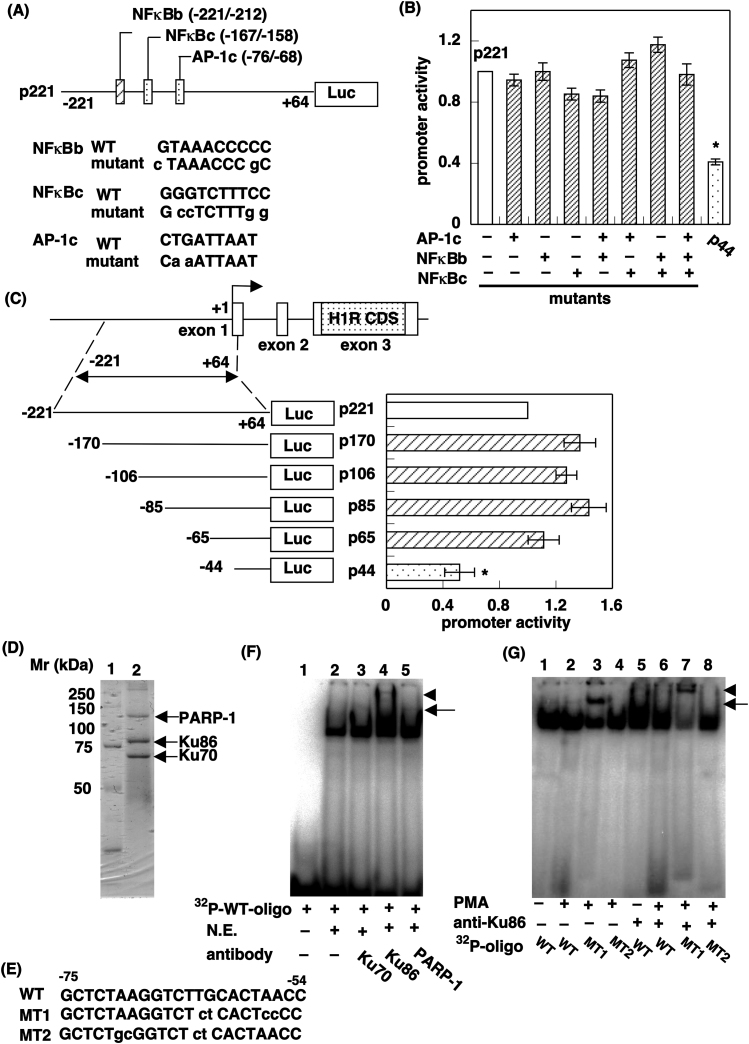
Figure 3. Identification of Ku86 as a protein that binds to region B1 of the promoter.
(A) Upper panel: Schematic diagram of region B, which contains two putative NF-κB binding sites (NF-κBb and NF-κBc) and one putative Ap-1 binding site (AP-1c). Mutations are indicated by lowercase letters. Lower panel: Nucleotide sequences of WT and MT oligonucleotides for the three putative transcription factor binding sites. (B) Luciferase reporter assay. Data are presented as mean ± S.E.M. (n = 9). *, p < 0.01 vs p221. (C) Identification of the minimum sequence responsible for promoter activity in region B. Data are presented as mean ± S.E.M. (n = 8). *, p < 0.01 vs p221. (D) DNA affinity chromatography. The nuclear extract (5 mg protein) was incubated with annealed biotinylated oligonucleotides bearing the region B1 for 30 min at 25°C. Beads were collected and boiled with 1× SDS-sample buffer. Proteins specifically bound to DNA were visualized by Coomassie blue R250 and identified by MS/MS. (E) Nucleotide sequences for oligonucleotides of region B1. Mutations are indicated by lowercase letters. (F and G) EMSA. The nuclear extract was incubated with 32P-labeled WT probe bearing the WT oligonucleotide sequence. For the supershift assay, the nuclear extract was incubated with 32P-labeled WT probe with anti-Ku70, anti-Ku86, or anti-PARP-1 antibody. In G, the nuclear extract prepared from HeLa cells with or without PMA stimulation was incubated with 32P-labeled WT (lanes 1, 2, 5, and 6), MT1 (lanes 3 and 7), or MT2 (lanes 4 and 8) probes. For the supershift assay, anti-Ku86 antibody was used. The positions of the sequence-specific DNA–protein complex and the shifted band are shown as an arrow and arrowhead, respectively.