Abstract
This Perspectives is a review of the breathtaking history of mammalian genetics in the past century and, in particular, of the ways in which genetic thinking has illuminated aspects of mouse development. To illustrate the power of that thinking, selected hypothesis-driven experiments and technical advances are discussed. Also included in this account are the beginnings of mouse genetics at the Bussey Institute, Columbia University, and The Jackson Laboratory and a retrospective discussion of one of the classic problems in developmental genetics, the T/t complex and its genetic enigmas.
THE field of genetics experienced explosive growth in the 20th century. It traveled from the rediscovery of Mendel in 1900, to the discovery of DNA as the heredity material, and, for a finale, to the sequencing of the human genome in 2001. In the last four decades of the 20th century, the pace of discovery was such that it seemed one could hardly catch one’s breath. In a parallel time frame, developmental biology morphed from a descriptive science into developmental genetics and molecular biology. A capsule history of the major discoveries in pure genetics and in mammalian/developmental genetics is presented in Table 1. Table 1 is by no means complete but is meant to be a personal overview and comparison time line of pure genetics verses mammalian developmental genetics.
Table 1 . Parallel time lines that trace the history of genetics and mammalian developmental genetics.
| Genetics | Mouse and developmental genetics | ||
|---|---|---|---|
| Scientist(s) | Contribution | Scientist(s) | Contribution |
| 1850: Darwin | Evolution | 1850–1940s: Hans Spemann and the German school | Long history of descriptive and experimental embryolgy |
| 1866: Mendel | Laws of inheritance | ||
| 1900: Hugo de Vries, Carl Correns, and Erich von Tschermak | Rediscovery of Mendel | 1900: Miss Abbie Lathrop | Starts breeding “fancy mice” |
| 1906: William Castle and The Busey Institute | Founder of mouse genetics | ||
| 1910s: Thomas Hunt Morgan | Sex-linked inheritance, gene theory, principle of linkage, chromosome theory of inheritance | 1908–1920: C. C. Little, Leonell Strong | Development of inbred stains |
| 1927: H. J. Muller | X rays are mutagenic in Drosophila | 1927: Dobrovolskaia-Zavadskaia | Discovery of Brachury, the first developmental mutation in mice and its interaction with t alleles |
| 1928: Griffith | Transfer of DNA from one bacterium to another | 1929: C. C. Little | Founded The Jackson Laboratory |
| 1931: Barbara McClintock | Recombination is caused by a physical exchange of chromosomal pieces | 1930: An academically inbred group of geneticists | 31 loci and 7 linkage goups defined |
| 1933: L. C. Dunn and Salome Glueckshon-Schoenheimer | Effectively found developmental genetics with the study of the T/t-complex | ||
| 1935: Elizabeth Fekete | First successful transfer of fertilized ova | ||
| 1937: George Beadle and Boris Ephrussi | Foundation for one gene/one enzyme hypothesis | 1936: The Jackson Laboratory researchers | First link between cancer and viruses in mammals |
| 1941: George Beadle and Edward Tatum | One gene/one enzyme hypothesis | 1942–1948: George Snell | Develops congenic strains of mice—identical but for a small chromosomal segment—by breeding for differences only at the H2 locus. This opens new areas of immunogenetics. |
| 1944: Avery, McCloud, and Macarty | Nucleic acid is the vehicle of heredity | ||
| 1944: Barbara McClintock | Hypothesis of transposable elements to explain color variations in corn. | ||
| 1951: Rosalind Franklin | Obtained sharp X-ray diffraction photographs of DNA. | 1950: Margaret Dickie | Obese mouse is discovered. The first animal model for obesity, the mouse later proves to have a mutation key in identifying the leptin gene. |
| 1952: Alfred Hershey and Martha Chase | DNA rather than proteins carries genetic information | 1953–1956: L. B. Russell | Dominant white spotting and steel loci identified |
| 1953: Watson and Crick | Resolved the structure of DNA | 1954: Leroy Stevens | Biology of stem cells in teratocarcinomas in an inbred strain |
| 1956: Vernon Ingram | A single amino acid difference in a protein (hemoglobin) can cause a disease | 1958: Margret Green | Database of mouse linkages and loci, which forms the foundation of the Mouse Genome Database |
| 1960s: Susumu Ohno | The total amount of chromosomal material in mammals was the same. Mammalian X chromosomes are conserved among species. | ||
| 1961: F. Jacob and J. Monod | The operon | 1961: Mary Lyon | X inactivation, the Lyon Hypothesis |
| 1960s: Tarkowski and Mintz | Mouse embryonic chimeras | ||
| 1970: H. Temin and D. Baltimore | Reverse transcriptase | 1970: Elizabeth Russell and T Mayer. | Steel(Kit) and dominant white spotting (W). pioneers the use of bone marrow transplantation to cure a blood disorder in a mouse. |
| 1970: Hamilton O. Smith | Restriction enzymes | 1971: Alfred Knudson | Genetics of retinoblastoma, loss of heterozygosity |
| 1971: Don Bailey and Ben Taylor | Recombinant inbred strains used for mapping | ||
| 1972: M. N. Nesbitt and U. Francke | Chromosomal banding and the first official idiogram of the mouse was produced | ||
| 1976: M. Biship and H. Varmus | Oncogenes | 1973: Susumu Ohno | Conservation of synteny; bits of linkages among mammels are conserved. |
| 1977: P. Sharp and R. Roberts | Splicing | 1975: D. Solter and B. Knowles | Immunosurgery of mouse blastocyst |
| 1977: Sanger and Maxam and Gilbert | DNA sequencing | 1980: C. Nüsslein-Volhard, E. Wieschaus, and E. B. Lewis | Screen to identify a set of genes crucial for Drosophila embryogenesis |
| 1981: Mario R. Capecchi, Martin J. Evans, and Oliver Smithies | ES cells were first independently derived from mouse embryos by two groups who also showed that ES cells could contribute to an embryo to form a chimera. | ||
| 1982: Richard Palmiter and Ralph Brinster | Transgenic mice | ||
| 1983: Kary Mullis | Invented the polymerase chain reaction | 1992 : F. Bonhomme and J.-L. Guénet | Outcross to Mus subspecies to gain polymorphisms for mapping |
| 1984: Vernon Bode | ENU mutation at specific loci in the t-complex | ||
| 1983 : Nancy S. Wexler et al. | Huntington’s is first human disease to be genetically mapped | 1984: Surani and McGrath and Solter and Cattanach and Kirk | Imprinting |
| 1984: McGinnis et al. | Discovers that homeotic (Hox) regulatory genes, responsible for the basic body plan of most animals, are conserved from flies to mammals | ||
| 1985: A. J. Jeffreys | Use of mini/microsatellites for mapping and forensics | 1987: Mario R. Capecchi, Martin Evans, and Oliver Smithies | First knockout mouse |
| 1989: Francis Collins and Lap-Chee Tsui | Identified the gene causing cystic fibrosis | ||
| 1990: Various | Launch of the Human Genome Project | 1990: B. Herrmann et al. | Positionally cloned T |
| 1993: Victor Ambros et al. | Identified microRNAs in Caenorhabditis elegans | 1996: Ian Wilemut | First cloning of a mammal in sheep |
| 1997: R. Yanagimachi | First cloning of a mouse from a somatic cell | ||
| 2001: Francis Collins and Craig Venter | Sequence of the human genome | 2002: Mouse Genome Sequencing Consortium | Sequence of the mouse genome |
| 2000–present “Age of omics” | 2000–present: Systems biology | ||
| 2006–2007 | Pluripotent stem cells artificially derived from a mouse adult somatic cell by inducing a “forced” expression of specific genes | ||
This table includes a personal but not comprehensive choice of high points in genetics and in mammalian developmental genetics.
The progress, however, was never steady; there were both stumbles and rapid accelerations. At midcentury, in particular, the separate fields of developmental genetics and embryology were running out of questions to ask that could be addressed with the tools at hand. And, by 1980, they had more new genetic markers and developmental mutations than this rather small and intellectually inbred group could handle. From roughly that point onward, geneticists had to absorb the thinking and master the techniques of the new molecular biology, a total change of paradigm.
This Perspectives is about 20th century genetics from the standpoint of mouse/mammalian genetics, a mouse geneticist’s eye view. To bring the reader through this exciting time, following the changing thread of ideas, I will use examples of interesting genes and present some of the high points of mammalian genetics in this period. They all presented puzzles that my generation had to cut their teeth on. There is no attempt to cover all of the relevant early experiments, and only a small selection of the high points will be discussed. The author apologizes to all whose discoveries are not included.
Why study the history of genetics? What seems so difficult for a modern student to envision is how each experiment fit into the framework of what was known at the time, given the inevitable gaps and errors in understanding and the seemingly limited nature of the techniques that were available. In retrospect, it all seems so obvious once there are no intellectual or technical barriers. Thus, if today’s geneticists engaged in data gathering and “mining” never grasp the ingenuity or technical fearlessness of the history of the field, they are doomed never to experience those qualities in a modern context. If we do not continue to employ hypothesis-driven research, genetics will lose much of its intellectual power. The other loss will be that of the courage to believe that proving a hypothesis is doable, even in the face of widespread disbelief.
My favorite example of hypothesis-driven research is an experiment of Susumo Ohno (Atkin et al. 1965). He was interested in the function and evolution of chromosomes, and thus of genetic material. In particular, he was fascinated by mammalian chromosomes. In the 1960s, after observing many mitotic figures in different mammalian species, Ohno had the impression that, even though each species had a different number of chromosomes and different chromosome morphologies, he was looking at the same amount of genetic material in the different species. How could you test this idea before the advent of molecular biology? What Ohno did was to photograph the mitotic preparations from several species under very carefully controlled conditions, then cut out the chromosomes from the photographic print of single-cell chromosome spreads, and finally weigh each cell set on a balance, comparing the results for the different species. Thus he was able to demonstrate that all the species included had the same amount of genetic material. It would take another five decades—until the genomes of several mammalian species had been sequenced—to confirm that he was correct.
Thus, a history of genetics must be appreciated in the conceptual framework of the time. As this might seem obscure to a student of 21st century genetics, I will attempt to describe that context as we go along. Much of the history of early mammalian genetics, when one studied one gene at a time, was nevertheless grounded in a thorough knowledge of biology. This foundation seems in danger of being lost with today’s emphasis on molecular biology, the various “omics,” and systems biology approaches. Yet the knowledge of the actual biology of organisms remains just as essential today.
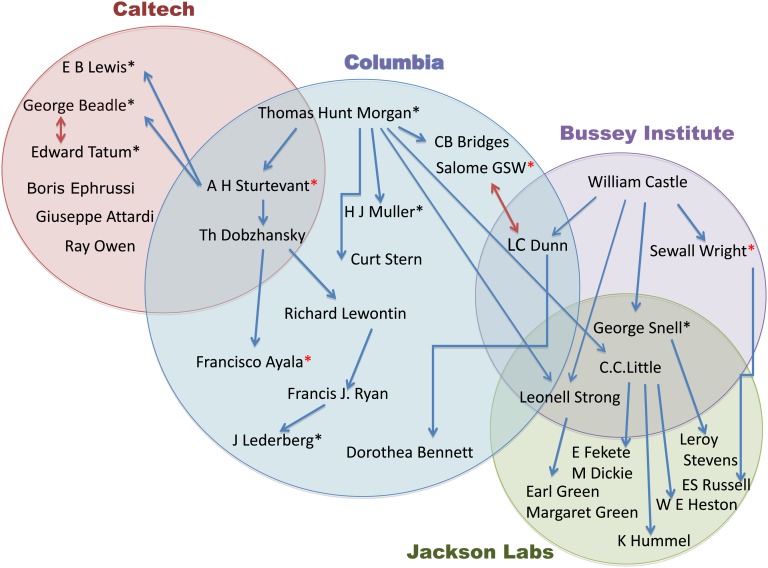
There were three great institutions that served as incubators of genetics and mammalian genetics in the first half of the 20th century: Columbia University, starting with Thomas Hunt Morgan’s fly genetics in the 1910s and continuing with mice through the 1950s; The Bussey Institute at Harvard that existed from 1908 to 1936 started with William Castle and his many famous students; and finally, The Jackson Laboratory (http://www.jax.org/milestones/researchhighlights.html) founded by C. C. Little in 1929. Those institutions and their scientific personnel (see Figure 1) compose vital elements of the story. Figure 1 is an attempt to show the historical overlap between institutions, pioneering researchers, and thus ideas. The arrows indicate mentors and students.
Figure 1 .
Institutions that served as incubators of genetics and mammalian genetics in the first half of the 20th century. A name within a circle or in overlapping circles shows that the scientist was present at that institution at a critical time. A blue arrow points to a student or post-doc, a red double arrow indicates collaborators, a single asterisk indicates a Nobel Prize winner, a red asterisk indicates a National Medal of Science winner. George Snell was also a post-doc of H. J. Muller.
T. H. Morgan’s Fly Room: The Beginning of Modern Genetics
No history of mammalian genetics can be written without first paying homage to the early Drosophila geneticists at Columbia (Figure 1). Starting with Thomas Hunt Morgan and his student, A. H. Sturtevant, the Drosophilists first had to identify mutations. Morgan’s earliest mutant, white eye (now designated simply white), led him to hypothesize sex linkage. And from there and other sex-linked mutations, they determined that genes were arranged on chromosomes in a linear fashion, the so-called “beads on a string” hypothesis. They also showed that the gene for any specific trait was at a fixed locus and the important concept that the frequency of crossing-over between two genes could determine their relative proximity on a linear genetic map.
Morgan’s fly room at Columbia became world famous, and the center of an informal exchange network, through which promising mutant Drosophila strains were transferred freely from lab to lab. In 1928, however, Morgan’s group moved to the California Institute of Technology where they continued to be an epicenter of genetics, attracting young scientists such as George Beadle, Boris Ephrussi, and Edward L. Tatum, who would soon become leaders in the field.
Columbia’s loss of the fly geneticists left a momentary vacuum, into which stepped the mouse geneticists, in particular William Castle’s student L. C. Dunn. In 1914, Dunn had hoped to be an undergraduate researcher in Morgan’s “fly room,” but, as he put it in 1965 in his Short History of Genetics, “as I stood in the doorway … it was obvious that there was little room.” Later, in 1928, when Dunn returned to the Zoology Department at Columbia as a professor, it fell to him to convert the fly room to one suitable to working with mice. Yet, preparing for that task was instructive in itself. “The ingenuity with which simple homemade installations had been made to serve as the basis for major scientific advances became apparent to me” (Dunn 1965).
William Castle and The Bussey Institute
After the rediscovery of Mendel’s work, William Castle became the father of mammalian genetics. Like the fly geneticists before him, however, he first had to find and identify mutations. In 1903, Castle showed that the albino phenotype was caused by a Mendelian recessive gene (see Morse 1978). Later, in 1909, with J. C. Phillips, he transplanted the ovaries of a black animal into an albino female and mated her to an albino male. All of the many offspring were black. Thus, in one ingenious experiment, two important ideas were proved: the germ cells are separate from the somatic cells, and genes are not contaminated by the environment (Morse 1985, page 113). How simple, how elegant a way this was of nailing two key principles at once.
In 1908, Castle became the director of The Bussey Institute at Harvard, the now famous incubator of mouse geneticists. Clarence C. Little, who was later to become the founder of The Jackson Laboratory, soon joined Castle in his studies. Together they established that the pink-eyed dilution (p) locus as separate from dilute (d) and albino (c) and showed that dominant yellow (Ay) was lethal in homozygotes (Russell 1978). This work served as the foundation for the first demonstration of linkage in the mouse.
By 1917, the groundwork for most subsequent studies in mammalian genetics had been laid through the work of Castle and his students. An important aspect of The Bussey Institute was Castle’s philosophy of collegial interaction. “Active discussions of problems under study were greatly encouraged in daily exchanges and in the weekly seminars in which both staff and students participated. Differences in opinion were freely aired but kept impersonal, and Castle was remarkably tolerant of students who voiced opinions contrary to his beliefs. In those instances when his own work or that of his students showed him to be wrong, he would publicly confess the error and provide the correct solution” (Morse 1985). Interestingly enough, these productive attitudes pervaded the laboratories of not only Castle’s students and his student’s students, but in some cases, their students, well into the 1950s–1970s.
Over a period of 28 years, The Bussey Institute trained 49 students, including L. C. Dunn, Clarence Little, Sewall Wright, Lionel Strong, and George Snell. Most students of mouse genetics can trace their scientific heritage back to Castle in one way or another (see figure 4 in Morse 1978).
Inbred Strains of Mice
A major contribution of Castle’s group, and of Clarence Little in particular, was the realization of the need for and development of inbred, genetically homogeneous lines of mice, a lesson well learned from Mendel. The first mating to produce an inbred line was begun by Little in 1909 and resulted in the DBA strain. By 1918, Little developed some of the most famous inbred lines that are still used today (Russell 1978).
An inbred strain is constructed by brother–sister or parent–offspring matings for a minimum of 20 generations. At that point, the mice are skin-transplant compatible and genetically identical. Inbred lines have played a crucial role in all areas of mouse genetics by allowing independent researchers to perform experiments on the same genetic material, which in turn allows the direct comparison of any results obtained in different laboratories around the world.
Curiously enough many of the inbred strains started not with professional researchers but with a hobbyist, Abbie Lathrop, who was the first American mouse “fancier.” She was a retired schoolteacher who around 1900 began to breed mice for sale as pets from her home. She reputedly began with a pair of waltzing mice but was so successful that she soon found herself in the small animal pet business. Her biggest clients, however, turned out to be the “new” mouse genetics researchers at the nearby Bussey Institute and other research institutions (Morse 1978). Together, they managed to turn curiosities into experimental material.
The Jackson Laboratory
Another crucial contribution of Little to mouse genetics was the role that he played in founding The Jackson Laboratory, also now known as JAX, in Bar Harbor, Maine. The laboratory was inaugurated in 1929 as the “natural heir” to The Bussey Institute (Snell and Reed 1993). Importantly, Little also served as its first director for 27 years.
The impetus for The Jackson Laboratory’s founding was to focus on mouse genetics and to use inbred strains for cancer research. Those strains were soon made widely available to other laboratories. Yet not all was plain sailing; the laboratory had to weather extreme difficulties early in its history. First, the stock market crash that occurred soon after it was founded made funding difficult right from the start. That funding was obtained is a testimony to the charisma of Little. The second problem encountered, in fact a disaster, was that in 1947 a drastic fire occurred that destroyed most of Mount Desert Island, including the main laboratory, which resulted in the loss of almost every mouse. That was when the benefits of having provided inbred mice to other investigators saved mouse genetics. From everywhere, there came offers of inbred stocks to help the laboratory get started again. Soon, almost all had been reconstituted and JAX was back in business.
JAX was the site of many achievements in mammalian genetics. A list of the tip of the iceberg would include: George Snell’s discovery of the genetic factors (those of the major histocompatibility complex, Snell 1948) that determine transplantation of tissue from one individual to another; the first evidence that viruses can cause cancer (Jackson and Little 1933); the discovery of the existence and nature of stem cells (Stevens 1958); the first successful mammalian bone marrow transplantation to cure a blood disorder (Lawson et al. 1956); and parabiosis experiments that led to the discovery of the hormone leptin (Coleman, and Hummel 1969). Beyond those discoveries, perhaps JAX’s great contribution was that, to many young investigators and summer students, it was a magical place. What made it so was the combination of its isolation, its beautiful setting, the chance to work with an unlimited number of mice, the variety of mouse mutants, and its very special welcoming attitude for young investigators. It became a mecca for mouse geneticists and provided a model for the formation of research institutes elsewhere.
Developmental genetics: L. C. Dunn and the Tt complex
A sea change in thinking occurred with the marriage of development biology and genetics. Many people today regard this as something that occurred in the 1980s, but it was, in fact, an intellectual fusion of disciplines that had started in the early 1930s at Columbia with L. C. Dunn and his collaborator Salome Glueckshon-Schoenheimer, a German–Jewish refugee who had trained in Hans Spemann’s embryology laboratory. Later in this endeavor, they were joined by Dorothea Bennett. These investigators, as shall be described in a moment, had the perfect genetic source materials at hand: mouse mutations at the so called “t complex.” Although the synthesis of developmental biology and genetics seems natural, indeed inevitable, today, it was certainly not always the case. Well into the 1980s there were university courses taught in developmental biology that emphasized descriptive events and embryo manipulations but ignored genetics. And certain high-profile molecular biologists, well into the 1970s, expressed skepticism about developmental genetics ever contributing anything.
Brachyury:
The genetics of what came to be called the “t complex,” located on chromosome 17, presented several violations of Mendel’s rules and was, in fact, the starting point of L. C. Dunn’s studies. The Brachyury (T) mutation was discovered in 1927 in a radiation experiment in the lab of Dobrovolskaia-Zavadskaia in Paris (Dobrovolskaia-Zavadskaia 1927). The original mutation was a substantial deletion. T is one of the very first developmental mutations known in mammals. It is semidominant, causing a short tail in heterozygotes and lethality in homozygotes. In the embryo, it is essential for the emergence and maintenance of mesoderm. T/T mice die shortly after gastrulation, displaying a complete loss of the posterior mesoderm due to primitive streak defects and the absence of a notochord. Ironically, half the amount of the wild-type protein made by this gene causes a seemingly trivial short tail, while absence of the protein, as occurs in homozygotes, produces embryo lethality for lack of an axis. Dobrovolskaia-Zavadskaia found that some wild-trapped mice carried a factor that interacted with T to cause taillessness. The first puzzle here was that a gene that had no ostensible phenotype on its own had an interaction with T. Dobrovolskaia-Zavadskaia referred to these as recessive t’s. She failed to make headway in dissecting the complexity of the t mutations that she found in wild mice, so she gave the mice to Dunn at Columbia University. “Dunny,” as he was called, and later Salome Glueckshon-Schoenheimer-Waelsch, were quick to latch on to the problem, but the secrets of this region of the genome were slow to reveal themselves. It posed several enigmas, which, in the end, took >70 years and the advent of molecular technology to sort out!
When T was finally cloned and identified in 1990 (Herrmann 1990), it was found to encode a novel transcription factor that is essential for mesoderm development. In fact, it contains a conserved domain (T box) that defined a whole new class of developmentally important transcription factors. There are presently 15 of them known to act at different points and in different tissues in development (Bollag et al. 1994; Naiche et al. 2005).
Tail interaction factor:
All wild-trapped t mutant chromosomes carry the same tail interaction factor (tct). It is now thought that T and tct are alleles, but the issue of allelism is still not entirely clear. Nevertheless, T acts as a diagnostic tool to determine the presence of t chromosomes in wild-caught mice because compound heterozygotes for T and tct (T/tct) cause taillessness. tct is a hypomorph because, on its own in homozygotes, it has no phenotype (see below). Allelism of T and tct was certainly not evident in the 1930s, so to be safe Dunny called them “pseudoalleles.” Yet, whatever the terminology used, it was not understood how mutations affecting tail length could be allelic with a series of different lethal mutations (see below) that affected the embryo in different tissues at different times. Later, these wild-trapped mutant chromosomes were called t-haplotypes, a term borrowed from immunogenetics and meaning a collection of linked genes that are inherited together as a group and symbolized by “tx,y.”
t-lethal mutations:
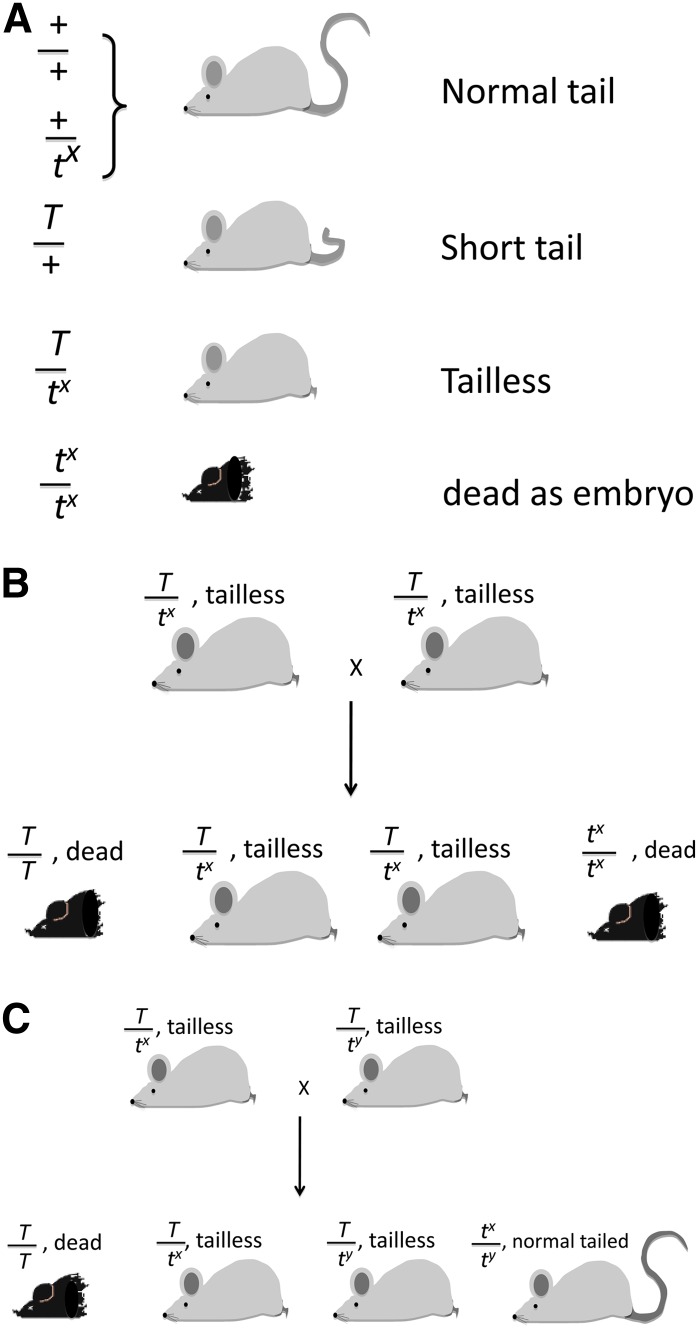
With rare exceptions, every t-haplotype trapped in the wild contains at least one recessive mutation lethal to the embryo. Some carry more than one as a hidden lethal (Paterniti et al. 1983). These would escape detection because, if homozygotes for one of the mutations died early, the death of a second one would be both undetectable and biologically irrelevant. The t-lethals were a series of different linked mutations defined by complementation between them. In this case, complementation meant that two tailless mice carrying the same mutation (T/tx) bred true for taillessness because both homozygotes (T/T and tx/tx) died, producing a balanced lethal system whereas, in contrast, two different ones (tx and ty) made one class of normal tailed progeny (Figure 2). In total, 16 different t complementation groups were identified (Klein et al. 1984). Dorothea Bennett studied many of the lethals extensively (Bennett 1964), finding that they affected different stages of development from morula to nearly completed fetus. These lethal or semilethal mutations were very informative for mammalian development. In fact, in the 1930s–1970s before large-scale mutagenesis and knockout technology were available, there were very few lethal mutations known. Virtually all of the then-known lethal mutations in the mouse were mapped to the t-complex.
Figure 2 .
Tail phenotypes and genotypes in Tt-complex crosses. (A) Tail phenotypes and genotypes are indicated. (B) The balanced lethal cross. It appears that taillessness breeds true because the other two genotypes die as embryos. (C) Complementation between t-lethals. Now a class of normal-tailed compounds tx/ty appears. The numbers of each phenotype will not resemble Mendelian ratios because the male transmits the t-haplotype to 99% of the progeny. The female has normal transmission. Thus one can expect ∼50% normal tails and 50% tailless offspring. Male normal tails are sterile. A variation of C is when the cross involves two parents carrying a semilethal (tsl). In this case, the phenotypes of the pups would be the same.
The rare exception of a t-haplotype carrying a gene that is not lethal is when some wild mice carry “semilethals.” This was a puzzling situation in which a proportion of homozygotes died as embryos, but the remainder of the litter developed normally with normal tails (Figure 2C). The normal-tailed mice are once again tct/tct but carrying two copies of t-semilethals. The existence of semilethals was a very puzzling phenomenon in the 1950s; this was before there was awareness of the effects of genetic background in mammalian development, although such effects were well known in Drosophila. Now we know, especially from knockout mice, that even a small amount of genetic background variation can make the difference between lethality and complete viability.
Recombination suppression:
Recombination suppression, as its name suggests, prevents genetic exchange. Its presence was soon demonstrated in crosses between t-haplotypes and wild-type (+) chromosomes. Whereas in wild type, T and an outside marker can be measured as 20 cM, the rate of recombination between t and wild type is only an exceptional 1/1000 or 0.1 cM. This was a genetic puzzle for half a century and was not completely understood until 1982. We now know that t-haplotypes contain four nonoverlapping inversions and behave like the balancer chromosomes known in Drosophila that also lock up an entire region. Sequencing has shown that the length of the t-complex region is >37 Mb and contains well over 500 genes, so the idea that it is a region of some unique developmental importance is no longer tenable. After the 1980s screens in Drosophila to identify genes crucial for embryogenesis (Jürgens et al. 1984; Nüsslein-Volhard et al. 1984; Wieschaus et al. 1984), it was clear that, in a region of this size in the mouse genome, many loci causing recessive embryonic lethality would be expected to occur. Nevertheless, as a result of this misconception of the unique importance of the t-complex region for development, and the additional fact that the major histocompatibility complex (H2) was included in the region, the consequence was that the proximal third of chromosome 17 became by far the best-mapped region of the mouse. This was true at a time when the rest of the mouse chromosome maps were still very sparsely populated with known mutations.
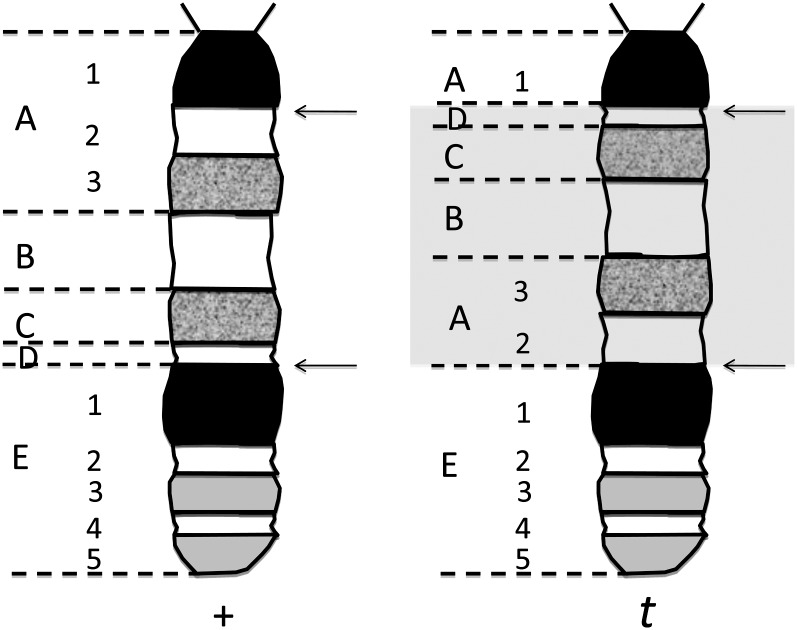
As noted above, we now know that the recombination suppression was due to four nonoverlapping inversions. In the mouse, these inversions, however, were cytologically invisible with the chromosome banding techniques available at the time. They all occurred between two distant light bands (Lyon, et al. 1988) (Figure 3). The 1/1000 exceptional recombinants mentioned above retained a mutant portion of chromosome 17 and gained wild type at the other end. Thus, when particular recombinants are viable, they can be detected as either normal tails (tct/tct) or by the exchange of an outside marker on the T chromosome.
Figure 3 .
The cytologically invisible inversions in a t-haplotype. The left chromosome is a normal one. The chromosome bands are indicated as they would have been seen with the techniques available in the 1970s. The inversions in a t-haplotype occur between bands A2 and D as indicated by arrows, highlighted with a light gray box. Without modern cytological techniques, these would not have been detectable. (This diagram has been modified from Lyon et al. 1988.)
These features argue that all t-haplotypes must be descended from a common ancestor (Silver 1993). Thus, for the most part, the whole t-haplotype travels genetically as a single unit.
Recombination suppression had been a nasty barrier to genetic analysis and understanding the t-complex. When Lee Silver and I solved the problem, we did not really have inversions in mind since the cytogeneticists had assured us that no inversions were involved. We just assumed that if “t-chromatin,” as Mary Lyon had dubbed it, was different from wild type and that if “good” chromatin would not recombine with “bad” (Lyon et al. 1979), then “bad” might recombine with “bad”!
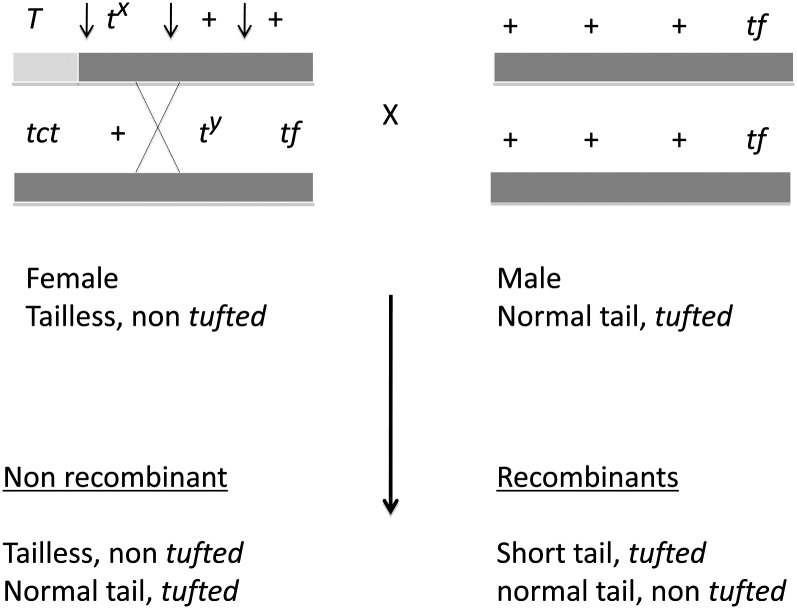
In 1981 (Silver and Artzt 1981), the experiment to test this could be done for two reasons. First, at some point in time, a spontaneous mutation to tufted (tf) occurred in a complete t-haplotype containing a lethal. tf is a hair-growth-pattern mutation ∼10 cM distal to T that can be visibly scored at 28 days after birth. Second, the T marker was available only in wild-type chromatin. However, in 1979, Mary Lyon showed that one of her t-haplotypes contained some “normal chromatin” at its very proximal end. Thus we were able to get recombination between it and T on a wild-type chromosome, effectively putting T together with distal t-chromatin. It was not until 1981 that Lee Silver and I configured the visible markers and two complementing lethals in a position where we could test recombination between two t-haplotypes.
The visible markers that we used were T and tf (Figure 4). Our first three litters contained 25% recombinant normal tailed (tufted) and short tailed (non-tufted). I remember returning to the mouse room several times that day to make sure that we had not mis-scored the mice for tf and thus our hypothesis was still true.
Figure 4 .
The cross that allowed measurement of recombination between t-haplotypes. Dark gray represents t-chromatin; light gray represents normal chromatin. The arrows represent the three possible intervals where recombination can be detected using the outside markers T and tf. Exactly which interval is involved must be determined by progeny testing for the lethals involved.
However, in 1981 before the inversions were known, this did not solve why t-chromatin could not recombine with wild type but, for the first time in 60 years, it gave us the tools to do recombinational genetics, map the different t-lethals, and solve the “pseudoallele problem.” Later studies showed that the recessive t-lethals were all non-allelic (Artzt et al. 1982).
Male germ-cell effects:
A final and most remarkable feature of t-haplotypes is transmission ratio distortion (TRD), which is found in male heterozygotes and the sterility found viable t homozygotes. Surprisingly, ∼10% of all mice trapped throughout the world contain a t-haplotype. This fascinated the population geneticists because, how could you explain the high frequency of such deleterious genes? TRD is common to all t-haplotypes and makes t’s look like a “viral epidemic” in terms of population genetics. In fact, it appeared that one t-haplotype (tw5) marched all the way from upper New York state to South America. The explanation is that any wild-derived t-haplotype is transmitted from heterozygous males (but not females) to upward of 99% of the progeny. In addition, a t-haplotype double heterozygote, as in complementing t-lethals (tx/ty), and t semilethal homozygotes are male sterile. In contrast, females of these genotypes transmit t-chromosomes in Mendelian ratios and are fertile.
The big question was, how do the TRD and male sterility of t-haplotypes work genetically? In 1979, Mary Lyon hypothesized that TRD involved interaction of three or more loci encoded in t-haplotypes. In her model, there were at least three or more segregation distorters and one responder element acting in cis, and these were all locked together by recombination suppression (Lyon et al. 1979). According to Lyon, the distorters were likely to be modules of dynein (Lyon 2003)
It became increasingly clear that Lyon’s model had been substantially correct. Nevertheless, the elements responsible for TRD were not unraveled for a decade or more, and there is still some disagreement about them. Although over time the number of putative distorters fluctuated, three of the distorter genes and the responder have been molecularly identified and all encode proteins that participate in RHO signaling pathways (Herrmann et al. 1999; Bauer et al. 2012).
In the 1970s, I wondered whether the genes for TRD were the same as those for sterility. Now it seems likely that they are not because, although many genes can cause sterility, even one of them locked up in recombination suppression and lethality can accumulate mutations without further consequences to the mouse (very much like the example of sterility and Qk below.)
steel and Dominant white Spotting
An intriguing biological puzzle for developmental geneticists in the 1960s was the steel (sl) and Dominant white (W) spotting mutations. Both are pleiotropic mutations with effects on pigment, hematopoiesis, and germ cells. In addition, both mutations have differing degrees of severity in heterozygotes and homozygotes; i.e., they are semidominant. The most interesting facet of their genetics, however, is that, although they reside on different chromosomes, they genetically interact.
Both mutations have been known for >60 years, and today there are a multitude of alleles at both loci, but the simple phenotypes that were known when both genes had just been identified from the initial alleles suffice to tell the tale. In most cases, surviving homozygotes for sl and W are black-eyed whites, very anemic and sterile, while heterozygotes for alleles of either one show a variable degree of color dilution and/or white spotting, anemia, and sterility. Compound heterozygotes as in sl/W essentially have the same phenotype as homozygotes for either gene. They are completely white with black eyes and are severely anemic or dead. This non-allelic, noncomplementation indicated that the W and sl gene products function in the same developmental pathway. What did these genes have in common except that all three affected cell types either had to migrate in the embryo or were derived from migrating cells?
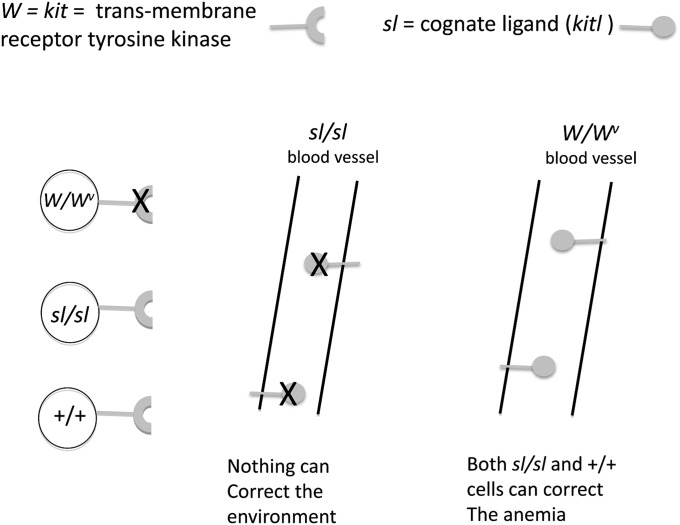
If one gene caused a cell defect and the other an environmental defect, an elegant and simple transplantation experiment done at The Jackson Laboratory in the 1960s would demonstrate it (McCulloch et al. 1965). It was found that steel mice cannot be cured of their hemapoietic defects with bone marrow cells from either W or wild-type mice, implying that steel has a pervasive effect on the microenvironment, preventing function or survival of the transplanted cells. In contrast, transplantation of W mice with bone marrow from either +/+ or steel mice resulted in a hematologic cure of the recipients via function of the transplanted cells (Figure 5). This result indicates that the defect in W mice is a defect intrinsic to the cell. Twenty years later, when both genes were cloned, this hypothesis was validated. How simple it is in retrospect: W, the first gene cloned, was the oncogene c-kit (also known as kit), which encodes a transmembrane tyrosine kinase receptor (Chabot et al. 1988; Geissler et al. 1988), and steel, the second to be cloned, was named its cognate ligand (kitl) (Anderson et al. 1990; Copeland et al. 1990).
Figure 5 .
Transplantation experiments with sl and W genotypes. Both mutants are severely anemic. In the case of sl/sl, the microenvironment is bad. Thus, no transplanted bone marrow, regardless of genotype, can survive. There is no ligand in the environment. In the case of W/Wv, the environment is good and those cells that have a normal c-kit (+/+ and sl/sl) can survive and rescue the mouse.
The multitude of spontaneous alleles of both genes are an added lesson in how genetics can help dissect biological and biochemical function because different alleles can affect different parts of the molecule and its interactions and, thus, have slightly different phenotypes. This was later shown to be of great usefulness when the mutagen N-ethyl-N-nitrosourea (ENU) was used to induce specific mutations (Bode 1984) and later when multiple alleles of many genes became available.
X Inactivation: The Lyon Hypothesis
Mary Lyon, R. A. Fisher’s last Ph.D. student, was both an “at the bench” researcher and an inspired theoretical geneticist. Working at the Medical Research Council Harwell in Great Britain with colleagues such as Charles Ford, Ted Evans, and Tony Searle, she was particularly interested in radiation biology and genetics, but some of her early work provided a landmark in the understanding of X chromosomes and mammalian sex differentiation. This work began with her interest in “calico” mice, which show a tricolor coat pattern much like the common calico cat. In 1960, she wondered why most X-linked mutants showed a variegated effect in heterozygous females and why the pattern of mottling resembled that seen in somatic mosaics. Her idea was that one X chromosome became inactive early in the development of a female embryo. She hypothesized that this happened at random so that either the maternally or the paternally derived X chromosome could be inactivated in different cells, but, once the choice was made, it was permanent. The resulting variegated coat color resulted from the mutant X chromosome that was active in some patches and the normal X chromosome that was active in others. Since males have only one X chromosome, it is always active. She further suggested that the inactive X chromosome was not only genetically inert but also heterochromatic. The latter idea would explain the long standing observation of the so-called Barr body, found only in the nuclei of female cells (reviewed in Lyon (2002).
Another important contribution of Mary Lyon that might seem unimaginably trivial today but which had a major impact is that, in the 1950s, she took over, from Margret Green at The Jackson Laboratory, an index card file of all mouse gene loci and mappings and incorporated these data into the Mouse News Letter or MNL. MNL was a “mimeographed” private publication circulated biannually and free to the community where mouse geneticists reported all their findings. This was the first mammalian genetics “journal,” and it functioned as such until Mammalian Genome incorporated MNL when it started publication in 1991!
Knockout Mice: Creation of Gene-Specific Mutations
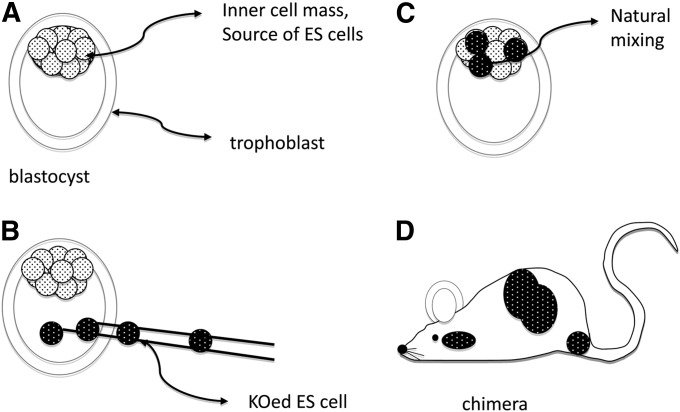
Probably the most important technical innovation of genetics in the 20th century was the ability to “knock out” (KO) specific genes by homologous recombination. This revolutionized mammalian genetics and biology. The first step involved the ability to establish cell lines from the uncommitted cells of the mouse embryo, the so-called embryonal stem cells (ES cells) (Evans 1981). In the beginning, manipulating the early mouse embryo faced several problems. At first it was the inaccessibility of the embryo itself that was a major technical hurdle. Once that was overcome, it was the inaccessibility of the inner cell mass. After superovulation, 2-day mouse embryos can be flushed from the oviduct. They are surrounded by an acellular shell that is easily removed by a limited protease digestion. At that point, the embryo is a very simple structure called a blastocyst (Figure 6). It consists of only two cell types. The inner cell mass that will give rise to the entire future embryo (these are the ES cells). But, the ES cells are rendered inaccessible because they are encased in the trophoblastic cells that will contribute to the extra-embryonic support tissues.
Figure 6 .
Making a knockout mouse. (A) Anatomy of the blastocyst. (B) KOed ES cells are injected into the blastocyst and (C) are allowed to mix naturally with the host inner cell mass cells. (D) After returning the embryos to a pseudopregnant female, some mice are born with patches of black from the manipulated cells.
The breakthrough in isolating viable and all important inner-cell mass cells was another example of an obvious idea that left all the developmental biologists knocking their heads and saying, “why didn’t I think of that!” In 1975, Davor Solter and Barbara Knowles invented a technique called “immunosurgery.” The used a variation of complement-dependent antibody cytotoxicity. Blastocysts were exposed to a rabbit anti-mouse antibody alone. After removing the antiserum, complement was added and only the trophoblastic cells were killed. The inner cell mass cells, never having been exposed to the antiserum, is healthy and can then be grown in culture (Solter and Knowles 1975). This method is still used extensively for deriving ES cells.
Once stable ES cell lines were established (Evans 1981), it was discovered that a construct containing two regions of homologous sequence flanking a deleted part of the gene could be electroporated into the ES cells (Smithies et al. 1985; Wong and Capecchi 1986). First, since homologous recombination is rather inefficient, a selectable marker for incorporation is necessary. Second, clones of treated cells need to be validated by Southern blot to assure that the recombination has in fact been correct. The construct inserted also contains a marker such as β-galactosidase so that the treated cells and gene can be visually followed.
What follows next is to turn the cloned cells into mice! It had been demonstrated much earlier that if you aggregate the uncommitted cells from two embryos together, the cells would intermingle and form a “chimera” (Buehr and McLaren 1974). Thus, if the two embryos were genetically destined to be two different coat colors, for example, black and albino, then the chimera would have patches of each color. Therefore, if a clone of homologously recombined cells was injected into the blastocyst, the ES cells would naturally mix with the inner cell mass and create a chimera.
However, there are still some genetics needed before one has a KO mouse. The progenitor cell presumably had only one copy KO’ed and is thus heterozygous (+/−). First, the chimeric male is mated to an albino. All the white offspring can be discarded, but the black offspring need to be tested for carrying the KO’ed allele. Two carriers are mated together to determine if −/− is viable or if there is a detectable mutant phenotype.
Homologous recombination, a classic genetic phenomenon, has thus proven to be part of the essential means by which genetic function in mice is currently studied. Indeed, there is a worldwide effort to knock out every mouse gene (Bradley et al. 2012).
quaking: How to Positionally Clone a “Deleted” Gene
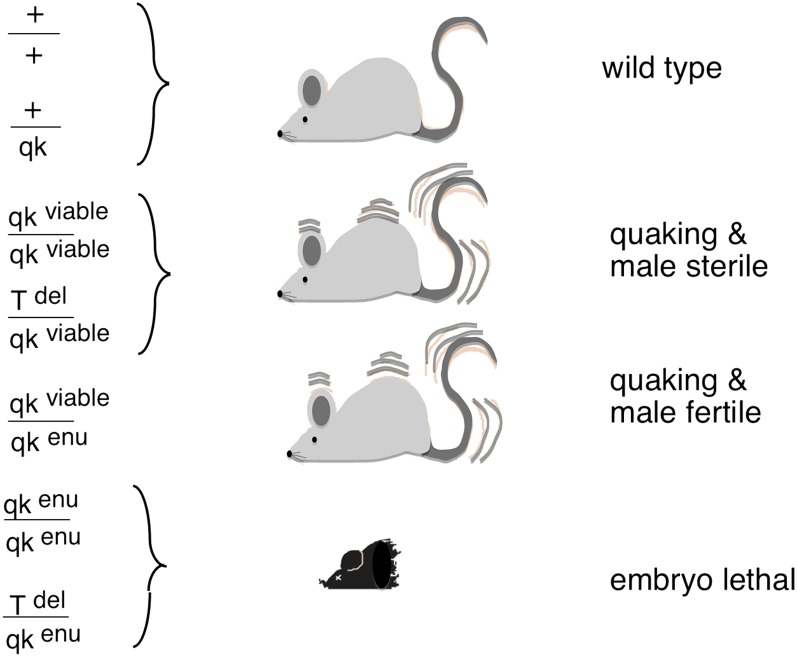
In the 1980s when positional cloning in mammals was becoming possible, it was a long and tedious task involving either cosmids (holding 40 kb of DNA) and chromosomal walking or, worse yet, tiresome microdissection of chromosomes. The latter procedure had been used for cloning T. My lab was interested in a pleiotropic mutation called quaking (Qk) that was included in the t-complex. Homozygosity for the original quaking mutation (Qkv) permitted viability but resulted in severe dysmyelination and male sterility. This phenotype contrasted with the other four existing, recessive, ENU-induced mutations that were all lethal at midgestation (Bode 1984; Justice and Bode 1988; Shedlovsky et al. 1988). The viable mutation was found to complement the lethality in Qkv/QkENU while the lethal ENU mutations complemented the Qkv male sterility. All of these compound heterozygotes, however, exhibited the quaking phenotype (Figure 7).
Figure 7 .
Phenotypes and genotypes of Qk mice.
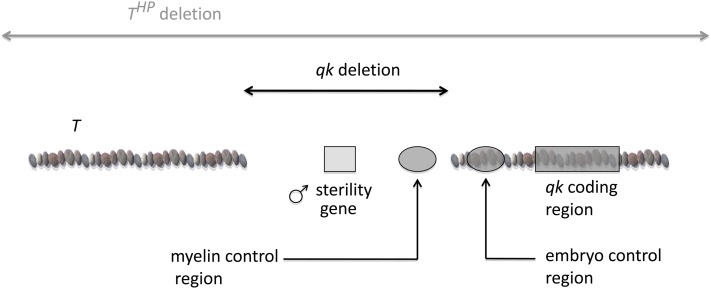
My lab was doing a considerable amount of mapping in the chromosomal region of Qk. Tom Ebersole, a graduate student in my lab, had managed to use whole cosmids as probes on Southern blots by blocking the repetitive sequences with total mouse DNA, a pretty gross technique in light of today’s sophisticated molecular protocols. To our surprise, a cosmid mapping to the Qk region and much of the surrounding DNA (at least 40 kb) was deleted (Ebersole et al. 1992)! This should have not been such a surprise because THP, a known, very large deletion around the locus of T, acted as a nullisomic for the nearby Qk locus. Thus, puzzling but forgotten by us was that THP/Qk was quaking. The whole story left us with the genetic riddle of how one could delete a gene that was necessary for embryogenesis yet still have viability. However, the historical context was that the literature was taking first note of tissue-specific enhancers, and this could in fact be part of the solution: a deleted tissue-specific enhancer preceding an intact gene.
The solution to identify a candidate Qkv gene had to be a purely genetic-based cloning strategy (Figure 8). As the four ENU-induced lethal alleles were presumably point mutations and Qkv complements all of them for their lethality but not their defective myelination, a single gene for these two functions of Qk was proposed. When the original Qkv mutation was found to be associated with a deletion, a model was introduced that predicted the location of the gene. Because the ENU mutations are recessive lethals, it is unlikely that a homozygous deletion of the Qk-coding sequences would be viable. The solution that we proposed was that two separate elements were necessary for Qk function and that the deletion interferes with a regulatory element necessary in the nervous system but not the embryo. Since regulatory regions were known to be generally close to coding sequences, the strategy was to clone the DNA flanking the deletion breakpoint. The qk gene was expected to be located very close to the breakpoint on one or the other side. In addition, since the compounds Qkv/QkENU were male fertile, a separate gene in the deletion must have been responsible for the male sterility (Figure 8).
Figure 8 .
The qk hypothesis. The deletion leaves the qk gene and its embryo enhancer intact but, removes an enhancer for the myelinating cells. The separate gene for male sterility is now known to be Pacrg (Lorenzetti et al. 2004).
As predicted, on one side of the breakpoint a candidate gene was found that was expressed in the earliest cells of the embryonic nervous system and in the myelinating tracts of prenatal brain. The hypothesis was correct (Ebersole et al. 1996)! The Qk gene turned out to be one of the founding members of a new family of RNA-binding proteins that act both in signal transduction and in RNA metabolism and that came to be called STAR proteins for signal transduction and activation of RNA (Vernet and Artzt 1997).
Conclusions
The beginnings of mouse genetics were strongly focused within a small group of scientists in the northeastern United States: first at The Bussey Institute, then at Columbia, and lastly at The Jackson Laboratory. At these institutions, the academic descendants of a very small number of key early figures in genetics were retesting Mendel’s principles in mice.
This article has tried to touch on examples of the several important paradigm changes that revolutionized mammalian genetics and some essential technical changes like the early advent of inbred strains of mice. Among the early experiments discussed was the dissection of development through the study of mouse mutations. A particular example was the work on the t-complex, a pioneering example of the use of genetic techniques to understand development. It thrust us into seeing the genetic underpinnings of mammalian development and developmental pathways.
An obvious, essential change was the transition from genetics to molecular biology. Consequently, one of the most important concepts to come out of 20th-century mammalian genetics was the slow realization that by descent from common ancestry the essential genes had been conserved though millennia. Starting early in the 1970s, the sequencing of genes from diverse species showed that parts of the essential genes were very similar. This meant that with some caveats, mice could be considered as a model organism to study the biology of these genes. It allowed developmental geneticists to do research on mice as a surrogate for humans and was responsible for a huge surge in mouse genetics.
Two final essential turning points that should be mentioned here are, first, the ability to manipulate the mouse embryo. This ultimately led to knockouts, whose development I have briefly reviewed here, and to the understanding of gene function. Finally, of course, there was the sequencing of whole genomes that now allows the relatively easy identification of genes and the hope of understanding complex biological systems.
Acknowledgments
I am grateful to Jiang I. Wu and Adam Wilkins for comments on this manuscript and to the many members of my laboratory for their inspiration.
Footnotes
Communicating editor: A. S. Wilkins
Literature Cited
- Anderson D. M., Lyman S. D., Baird A., Wignall J. M., Eisenman J., et al. , 1990. Molecular cloning of mast cell growth factor, a hematopoietin that is active in both membrane bound and soluble forms. Cell 63: 235–243 [DOI] [PubMed] [Google Scholar]
- Artzt K., McCormick P., Bennett D., 1982. Gene mapping within the T/t-complex of the mouse. I. t-Lethal genes are nonallelic. Cell 28: 463–470 [DOI] [PubMed] [Google Scholar]
- Atkin N. B., Mattinson G., Beçak W., Ohno S., 1965. The comparative DNA content of 19 species of placental mammals, reptiles and birds. Chromosoma 17: 1–10 [DOI] [PubMed] [Google Scholar]
- Bauer H., Schindler S, Charron Y., Willert J., Kusecek B., et al. , 2012. The nucleoside diphosphate kinase gene nme3 acts as quantitative trait locus promoting non-mendelian inheritance. PLoS Genet. 8: e100256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett D., 1964. Abnormalities associated a chromosome region in the mouse. II. Embryological effects of lethal alleles in the T-region. Science 144: 263–267 [PubMed] [Google Scholar]
- Bode V. C., 1984. Ethylnitrosourea mutagenesis and the isolation of mutant alleles for specific genes located in the T region of mouse chromosome 17. Genetics 108: 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag R. J., Siegfried Z., Cebra-Thomas J. A., Garvey N., Davison E. M., et al. , 1994. An ancient family of embryonically expressed mouse genes sharing a conserved protein motif with the T locus. Nat. Genet. 7: 383–389 [DOI] [PubMed] [Google Scholar]
- Bradley A., Anastassiadis K., Ayadi A., Battey J. F., Bell C., et al. , 2012. The mammalian gene function resource: the international knockout mouse consortium. Mamm. Genome 23: 580–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M., McLaren A., 1974. Size regulation in chimaeric mouse embryos. Embryol. Exp. Morph. 31: 229–234 [PubMed] [Google Scholar]
- Chabot B., Stephenson D. A., Chapman V. M., Besmer P., Bernstein A., 1988. The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature 335: 88–89 [DOI] [PubMed] [Google Scholar]
- Coleman D. L., Hummel K. P., 1969. Effects of parabiosis of normal with genetically diabetic mice. Am. J. Physiol. 217: 1298–1304 [DOI] [PubMed] [Google Scholar]
- Copeland N. G., Gilbert D. J., Cho B. C., Donovan P. J., Jenkins N. A., et al. , 1990. Mast cell growth factor maps near the steel locus on mouse chromosome 10 and is deleted in a number of steel alleles. Cell 63: 175–183 [DOI] [PubMed] [Google Scholar]
- Dobrovolskaia-Zavadskaia N., 1927. On the spontaneous death of the tail in newborn mice and the existence of a hereditary, non-viable character (factor). Crit. Rev. Soc. Biol. 97: 114–116 (in French). [Google Scholar]
- Dunn L. C., 1965. A Short History of Genetics. McGraw-Hill, New York [Google Scholar]
- Ebersole T. A., Rho O., Artzt K., 1992. The proximal end of mouse chromosome 17: new molecular markers identify a deletion associated with quakingviable. Genetics 131: 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole T. A., Chen Q., Justice M. J., Artzt K., 1996. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat. Genet. 12: 260–265 [DOI] [PubMed] [Google Scholar]
- Evans M. J., 1981. Origin of mouse embryonal carcinoma cells and the possibility of their direct isolation into tissue culture. J. Reprod. Fertil. 62: 625–631 [DOI] [PubMed] [Google Scholar]
- Geissler E. N., Ryan M. A., Housman D. E., 1988. The dominant-white spotting (w) locus of the mouse encodes the c-kit proto-oncogene. Cell 55: 185–192 [DOI] [PubMed] [Google Scholar]
- Herrmann B. G., Labeit S., Poustka A., King T., Lehrach H., 1990. Cloning of T gene required in mesoderm formation in the mouse. Nature 343: 617–622 [DOI] [PubMed] [Google Scholar]
- Herrmann B. G., Koschorz B., Wertz K., McLaughlin K. J., Kispert A., 1999. A protein kinase encoded by the t complex responder gene causes non-Mendelian inheritance. Nature 402: 141–146 [DOI] [PubMed] [Google Scholar]
- Jackson R. B., Little C. C., 1933. The existence of non-chromosomal influence in the incidence of mammary tumors in mice. Science 78: 465–466 [DOI] [PubMed] [Google Scholar]
- Jürgens G., Wieschaus E., Nüsslein-Volhard C., 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome Roux’s Arch. Dev. Biol. 193: 283–295 [DOI] [PubMed] [Google Scholar]
- Justice M. J., Bode V. C., 1988. Three ENU-induced alleles of the murine quaking locus are recessive embryonic lethal mutations. Genet. Res. 51: 95–102 [DOI] [PubMed] [Google Scholar]
- Klein J., Sipos P., Figueroa F., 1984. Polymorphism of t-complex genes in European wild mice. Genet. Res. 44: 39–46 [Google Scholar]
- Lawson F. A., Russell E. S., Smith L. J., 1956. Implantation of normal blood-forming tissue in radiated genetically anemic hosts. Science 124: 1076–1077 [DOI] [PubMed] [Google Scholar]
- Lorenzetti D., Bishop C. E., Justice M. J., 2004. Deletion of the Parkin coregulated gene causes male sterility in the quaking(viable) mouse mutant. Proc. Natl. Acad. Sci. USA 101: 8402–8407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M. F., 2002. A personal history of the mouse genome. Annu. Rev. Genomics Hum. Genet. 3: 1–16 [DOI] [PubMed] [Google Scholar]
- Lyon M. F., 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37: 393–408 [DOI] [PubMed] [Google Scholar]
- Lyon M. F., Evans E. P., Jarvis S. E., Sayers I., 1979. t-Haplotypes of the mouse may involve a change in intercalary DNA. Nature 279: 38–42 [DOI] [PubMed] [Google Scholar]
- Lyon M. F., Zenthon J., Evans E. P., Burtenshaw M. D., Willison K. R., 1988. Extent of the mouse t complex and its inversions shown by in situ hybridization. Immunogenetics 27: 375–382 [DOI] [PubMed] [Google Scholar]
- McCulloch E. A., Siminovitch L., Till J. E., Russell E. S., Bernstein S. E., 1965. The cellular basis of the genetically determined hemopoietic defect in anemic mice of genotype Sl-Sld. Blood 26: 399–410 [PubMed] [Google Scholar]
- Morse, H. C. (Editor), 1978 Historical perspective, Introduction in Origins of Inbred Mice, edited by H. C. Morse. Academic Press, New York ( http://www.informatics.jax.org/morsebook/)
- Morse H. C., 1985. The Bussey Institute and the early days of mammalian genetics. Immunogenetics 21: 109–116 [DOI] [PubMed] [Google Scholar]
- Naiche L. A., Harrelson Z., Kelly R. G., Papaioannou V. E., 2005. T-box genes in vertebrate development. Annu. Rev. Genet. 39: 219–239 Review [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E., Kluding H., 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I. Zygotic loci on the second chromosome Roux’s Arch. Dev. Biol. 193: 267–282 [DOI] [PubMed] [Google Scholar]
- Paterniti J. R., Jr, Brown W. V., Ginsberg H. N., Artzt K., 1983. Combined lipase deficiency (cld): a lethal mutation on chromosome 17 of the mouse. Science 221: 167–169 [DOI] [PubMed] [Google Scholar]
- Russell E. S., 1978. Origins and history of mouse inbred strains: contributions of Clarence Cook Little, Chapter 5 in Origins of Inbred Mice, edited by H. C. Morse. Academic Press, New York (http://www.informatics.jax.org/morsebook/)
- Shedlovsky A., King T. R., Dove W. F., 1988. Saturation germ line mutagenesis of the murine t region including a lethal allele at the quaking locus. Proc. Natl. Acad. Sci. USA 85: 180–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver L. M., 1993. The peculiar journey of a selfish chromosome: mouse t-haplotypes and meiotic drive. Trends Genet. 9: 250–255 [DOI] [PubMed] [Google Scholar]
- Silver L. M., Artzt K., 1981. Recombination suppression of mouse t-haplotypes due to chromatin mismatching. Nature 290: 68–70 [DOI] [PubMed] [Google Scholar]
- Solter D., Knowles B. B., 1975. Immunosurgery of mouse blastocyst. Proc. Natl. Acad. Sci. USA 72: 5099–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O., Gregg R. G., Boggs S. S., Koralewski M. A., Kucherlapati R. S., 1985. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature 317: 230–234 [DOI] [PubMed] [Google Scholar]
- Snell G. D., 1948. Methods for the study of histocompatibility genes. J. Genet. 49(2): 87–108 [DOI] [PubMed] [Google Scholar]
- Snell G. D., Reed S., 1993. William Ernest Castle, pioneer mammalian geneticist. Genetics 133: 751–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L. C., 1958. Studies on transplantable testicular teratomas of strain 129 mice. J. Natl. Cancer Inst. 20: 1257–1275 [DOI] [PubMed] [Google Scholar]
- Vernet C., Artzt K., 1997. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 13: 479–484 [DOI] [PubMed] [Google Scholar]
- Wieschaus E., Nüsslein-Volhard C., Jürgens G., 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. III. Zygotic loci on the X-chromosome and fourth chromosome Roux’s Arch. Dev. Biol. 193: 296–307 [DOI] [PubMed] [Google Scholar]
- Wong E. A., Capecchi M. R., 1986. Analysis of homologous recombination in cultured mammalian cells in transient expression and stable transformation assays. Somat. Cell Mol. Genet. 12: 63–72 [DOI] [PubMed] [Google Scholar]