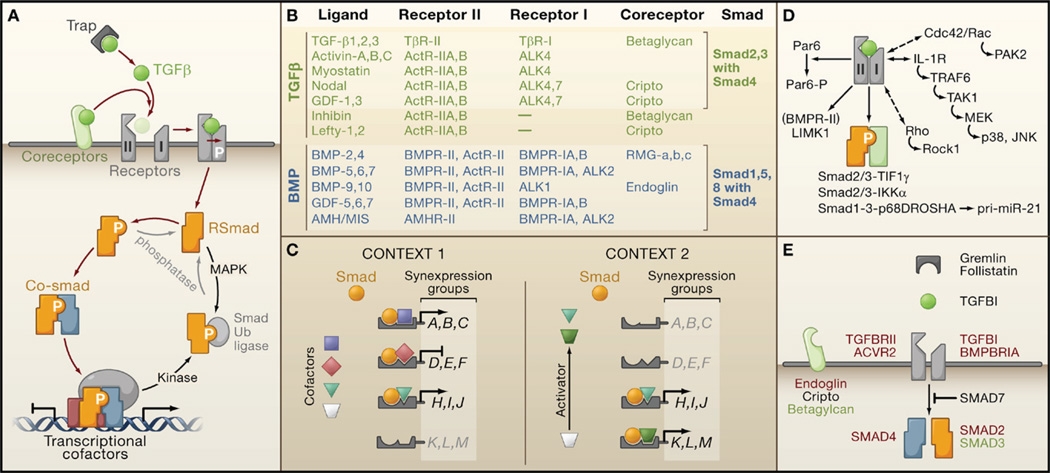
Figure 3. Organization of TGFβ Signaling and Weak Links in Cancer.
(A) Ligand traps and coreceptor molecules control the access of TGFβ family ligands to signaling receptors. The ligand assembles a tetrameric complex of receptor serine/threonine kinases types I and II. Receptor-II phosphorylates and activates receptor-I, which then phosphorylates and activates Smad transcription factors (RSmads). Activated RSmads bind Smad4 and further build transcriptional activation and repression complexes to control the expression of hundreds of target genes in a given cell. Mitogen-activated protein kinases (MAPK) and other protein kinases phosphorylate Smads for recognition by ubiquitin ligases and other mechanisms of inactivation. Phosphatases have been identified that reverse these phosphorylation events.
(B) An abridged chart of ligand-receptor-coreceptor-Smad relationships in the TGFβ (green) and BMP (blue) branches of the TGFβ family.
(C) Distinct combinations of transcription partner cofactors in different contexts (e.g., different cell types or conditions) determine the set of genes targeted by specific activated Smads. Each Smad-cofactor combination coordinately regulates a synexpression group of target genes. Smad signaling serves as a node for integrating regulatory signals that impinge on partner cofactors (e.g., Activator signal in Context 2).
(D) Alternative modes of TGFβ signaling include Smad4-independent RSmad signaling (via interactions with TIF1γ, IKKα, p68DROSHA), Smad-independent receptor-I signaling (via small G proteins and MAPK pathways), and direct receptor-II signaling (via Par6, and via LIMK1 in the case of BMPR-II).
(E) Core TGFβ pathway components that are affected by mutation (red), overexpression (black), or downregulation (green) in human cancers.