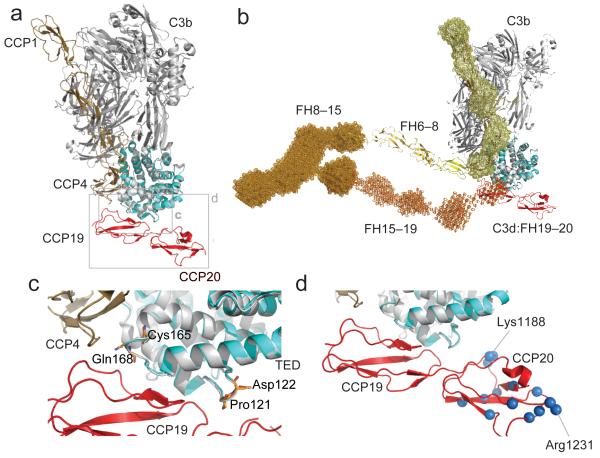
Figure 5.
Potential model of FH engagement with surface bound C3b (a) Superposition (C3d on TED) of the C3d:FH19–20 complex and C3b:FH1–4 complex demonstrating the close proximity of FH modules 4 and 19. (b) The FH19–20:C3d structure (red and cyan) is shown superimposed (C3d on TED) onto the FH1–4:C3b structure (tan and gray). This FH model has been constructed by juxtaposition or superposition of the structures of FH5 (yellow) and FH6–8 (yellow-orange) and the SAXS-shape envelopes of FH8-15 (bright orange) and FH15-19 (orange). Structures are shown in cartoon representation; SAXS shape envelopes are shown in mesh. Note, the FH1–4 SAXS shape envelope (pale yellow) is superimposed onto the C3b:FH1–4 structure. (c) Close up view of the α4-α5 loop and the α6-α7 loop of C3d. Highlighted in orange are the residues Pro121 (Pro1092), Asp122 (Asp1093), Cys165 (Cys1136) and Gln168 (Gln1139) for which disease-associated mutations have been reported. (d) Close-up view of the FH19–20 interaction with TED highlighting residues also like to interact with self-surface markers. Amides experiencing considerable chemical shift perturbations upon exposure to dp8 are highlighted as blue spheres (corresponding to their nitrogen atoms). Residues Lys1188 and Arg1231, which exhibit the greatest perturbations upon addition of dp8, are indicated.