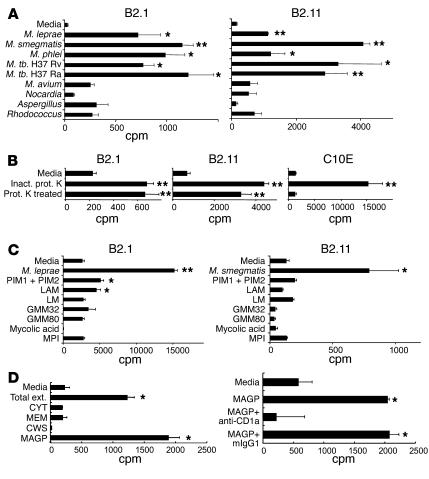
Figure 3.
Specificity of the proliferative responses of CD1a-restricted T cell clones to different bacterial lysates and known CD1-restricted antigens. (A) T cell lines were tested for their ability to recognize different total bacterial sonicates. M. tb., M. tuberculosis. (B) M. leprae sonicate preparation was digested with proteinase K. The proliferative response was determined using CD1a-restricted T cell line B2.1, B2.11 and the MHC class II–restricted cell line C10E. Inact. prot. K indicates that the enzyme was heat-inactivated prior to adding to bacterial extract. (C) Proliferative response was measured using different known CD1b- and CD1c-restricted antigens. Proliferative responses were measured as described for Figure 2 using LC-like DCs as APCs for T cell clones B2.1 and B2.11 and MHC-matched PBMCs for the C10E T cell line. PIM, phosphatidyl-myo-inositol mannoside; LAM, lipoarabinomannan; LM, lipomannan; GMM, glucose monomycolate; MPI, mannosyl phosphoisoprenoid. (D) Characterization of mycobacterial antigen for CD1a-restricted T cells. Left: CD1a-restricted T cell clone B2.11 was cultured with three mycobacterial extracts containing cytosol (CYT), membrane (MEM), soluble cell wall material (CWS), and mycolyl arabinogalactin peptidoglycan (MAGP). Total ext., mycobacterial extract prior to fractionation. Right: T cell–responsive MAGP was examined for CD1a-restriction using neutralizing antibodies to CD1a. The values shown are the mean ± SEM of triplicate cultures and are representative of at least three independent experiments. Statistical analysis was performed as indicated in the legend to Figure 2, but comparing T cells stimulated with media versus antigen. *P < 0.05; **P < 0.005.