Figure 2. SAHA, VPA, TSA, NaB, and DZNep alter the chromatin environment at the FIV promoter.
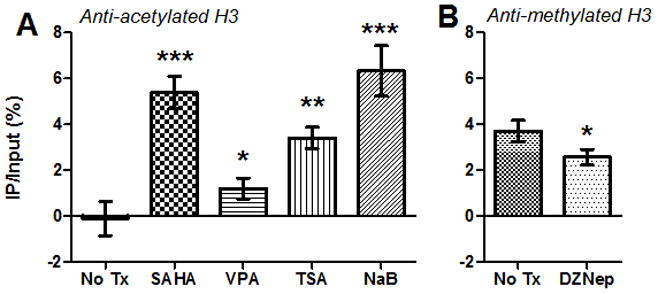
CD4+ T-cells from 2 asymptomatically FIV-infected cats were treated with HDACi (A), HMTi (B), or no treatment (No Tx), and chromatin immunoprecipitation (ChIP) assays were performed using anti-acetylated H3 (H3K9,14ac, Millipore, A) or anti-methylated H3 (H3K27me3, Millipore, B) antibodies. Following ChIP, quantitative PCR was used to detect a 180-bp fragment at the 5′ FIV LTR (forward primer GCTTCGAGGAGTCTCTCAGTTGAG at U3/R interface; reverse primer CAAATCAAGTCCCTGTTCGGGC in gag leader). Data are presented as the percentage of immunoprecipitated (IP) FIV DNA out of the total input FIV DNA with background subtraction using a normal IgG control. Error bars represent the standard deviation of triplicate qPCR measurements, and unpaired, one-tailed t-tests (GraphPad Prism, v5.04) were used to determine if there was a significant increase or decrease in histone acetylation or methylation, respectively (*p < 0.05, **p < 0.01, ***p < 0.001). Data are representative of two independent ChIP experiments for each condition.
