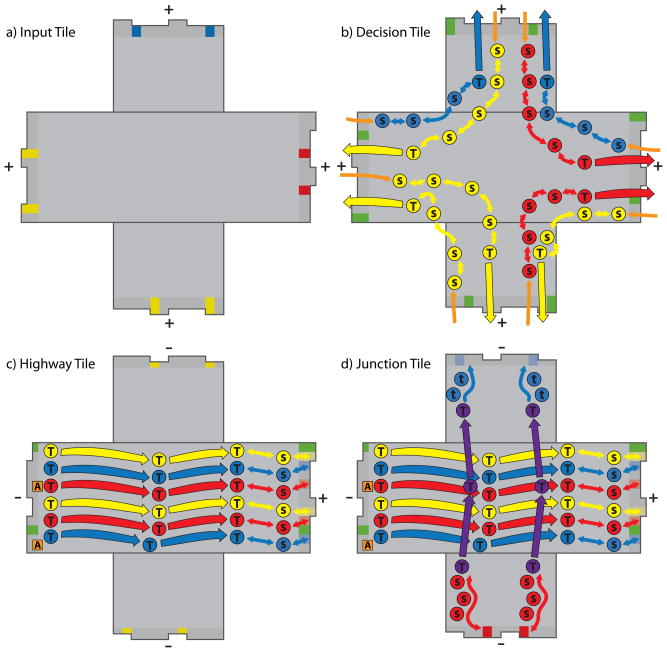
Figure 14.
Schematic representation of the four recursive Robinson tiles made from DNA and their signaling pathways. Binding sites (+,−), neutral seesaws (S), transducers (T,t), and activation strands (A) are indicated on each of the four tiles. Inactive binding sites are indicated with pale colors and two-way arrows between them and seesaw elements (S) on the Highway and Junction Tiles.. The direction of propagation of energetically favorable signal cascades is indicated with arrows. Seesaw pathways that can go in either direction are indicated with arrowheads in both directions. Positions of elements are based on the locations of free staple strand ends. a) The Input Tile contains only binding sites that are already active. b) Activation signals entering the Decision Tile from bound neighbors are indicated in orange. Each neighbor bound to the Decision Tile activates one of the signals on each of the adjacent edges. This arrangement means that two of the three other edges must be bound before a given edge of the Decision Tile can send its signal. This setup is much easier to implement than the 4-way AND gate that the logic of the tile system being implemented would nominally demand. This adjacent neighbor AND gate should suffice for practical purposes. Although there are only two unique activation strands, seesaw pathways (S) on the Decision Tiles are kept orthogonal by using orthogonal toeholds. c) Three different signals can propagate along the Highway Tile, ending in binding site activation as indicate by two-way arrows, or in the propagation of the signal directly from the last transducer (T) to the first transducer of a neighboring tile. d) The Junction Tile contains signaling pathways that cross one another. From left to right this tile has the same pathways as the Highway Tile. From bottom to top is the pathway for an red binding interaction to activate the blue binding site on the top. The orthogonality of this pathway is indicated by coloring the junction signal purple.