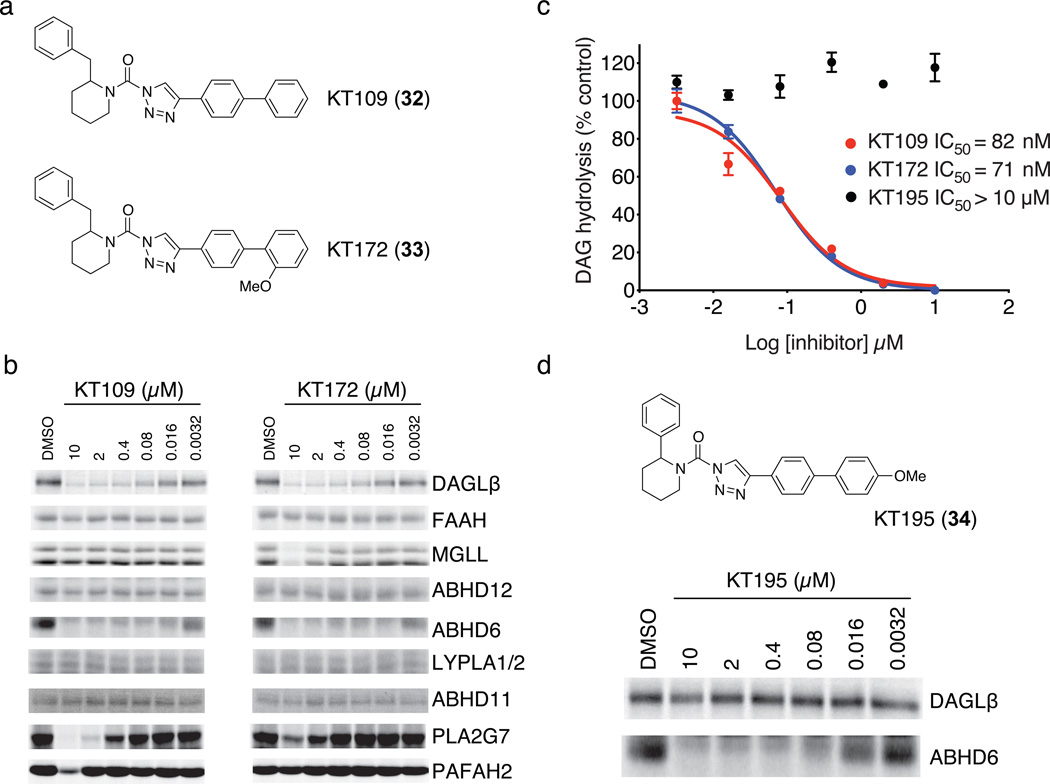
Figure 1. Optimization of 1,2,3-TU inhibitors for DAGLβ.
(a) Structures of optimized DAGLβ inhibitors KT109 and KT172. (b) Competitive ABPP of KT109 and KT172 against a panel of serine hydrolases detected in mouse brain proteome (FAAH, MGLL, ABHD12, ABHD6, LYPLA1 and LYPLA2 (LYPLA1/2)) or as enzymes recombinantly expressed in HEK293T cells (DAGLβ, ABHD11, PLA2G7, and PAFAH2). Proteomes were analyzed as described in Supplementary Fig. 3. See Supplementary Fig. 5 for IC50 values measured by competitive ABPP for KT109 and KT172 inhibition of DAGLβ. (c) In vitro IC50 values for DAGLβ inhibition by KT109 and KT172 measured with the SAG substrate assay following the protocol described in Supplementary Fig. 3c except SAG substrate was incubated with DAGLβ lysates for only 10 min at 37 °C after pretreatment with inhibitors. Data are mean ± s.e.m. for two independent experiments. 95% confidence intervals for IC50 values: KT109, 50–100 nM; KT172, 50–90 nM. (d) Structure and activity of control probe KT195. KT195 showed negligible cross-reactivity with recombinant DAGLβ (top) and concentration-dependent inhibition of ABHD6 (bottom) as measured by competitive ABPP.