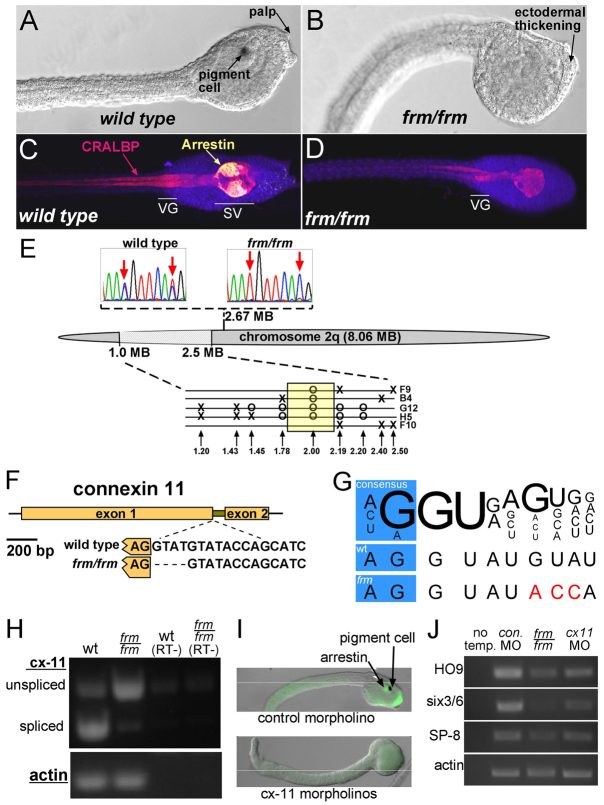
Fig. 1.
Characterization of the frimousse mutation. (A,B) Wild-type (A) and frm/frm (B) late tailbud stage Ciona intestinalis embryos. (C,D) Wild-type (C) and frm/frm (D) larvae immunostained for CRALBP (pan-CNS) and Arrestin (sensory vesicle). VG, visceral ganglion; SV, sensory vesicle. (E) Mapping of the frm mutation on chromosome 2q. Sequence traces (top) show linkage by SNP mapping to position 2.67 Mb. Finer mapping in the region 1.0 to 2.5 Mb with INDEL markers is shown (bottom). Single recombinant frm/frm tadpoles are indicated (F9, B4, G12, H5, F10). Circles indicate where only the linked INDEL allele was observed, and X indicates where a recombination resulted in both INDEL alleles being present. (F) The C. intestinalis connexin-like 11 (cx-11) gene, which in frm embryos contains a 4 bp deletion at the 5′ splice site of the single intron. (G) Comparison of the cx-11 5′ splice sites in wild type (wt) and in the frm allele with the consensus sequence (Wang and Burge, 2008). Variation from consensus are indicated in red. (H) RT-PCR for cx-11 using intron-spanning primers. Splicing is greatly reduced in cDNA from frm/frm embryos. RT–, controls without reverse transcriptase. (I) Injection of morpholinos (MOs) to cx-11 phenocopies the frm phenotype. (J) RT-PCR for three anterior neural plate-expressed genes (HO9, brain; six3/6, oral siphon primordium; SP-8, palp) and actin (loading control) in embryos injected with control MO or cx-11 MOs and in homozygous frm embryos (frm/frm). No temp., no template control.