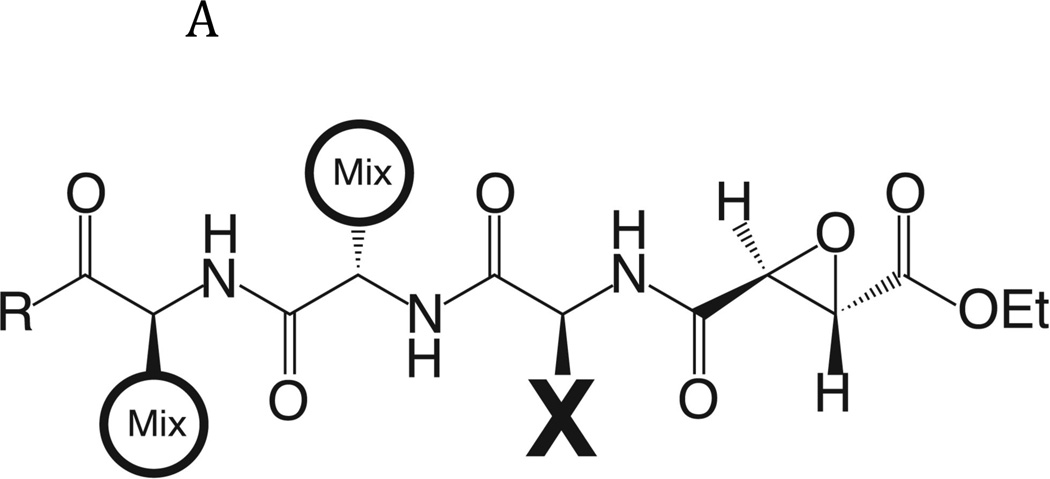
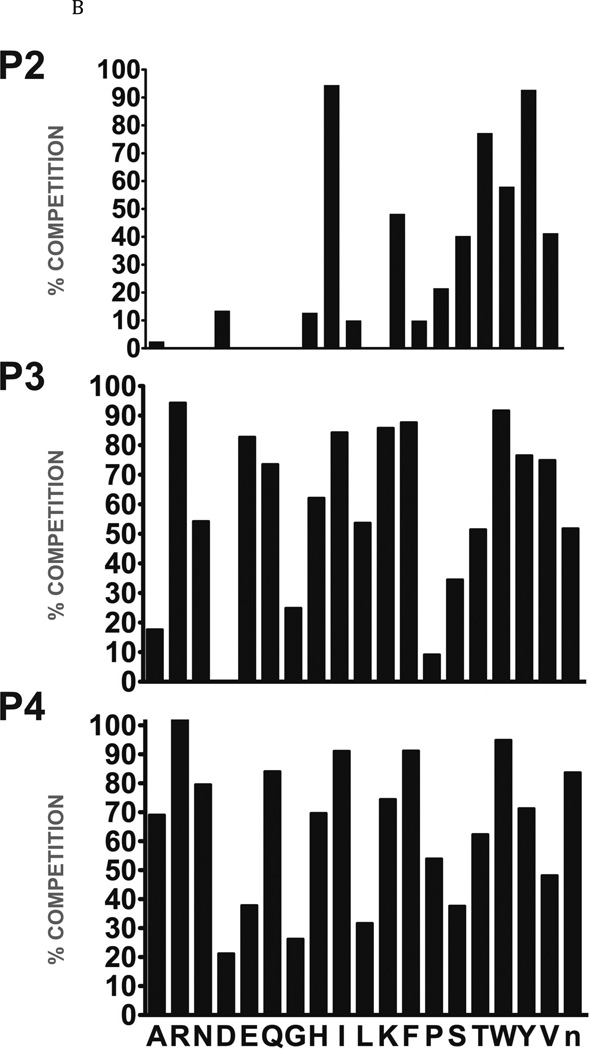
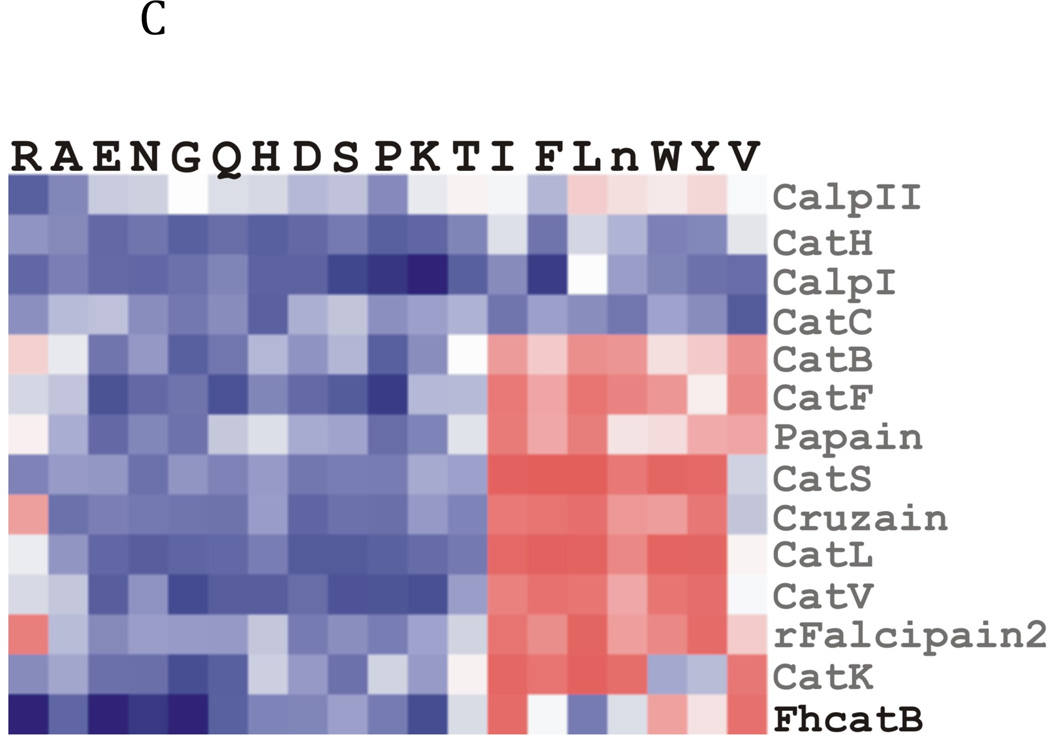
Fig. 2. Determination of the specificity of FhcatB1 using a combinatorial peptide inhibitor library.
A: Inhibitor scaffold used in libraries utilized to determine the specificity of FhcatB1. The scaffold encompasses a fixed position (indicated by ‘X’) which can be changed to each natural amino acid and norleucine, and two mixed positions (indicated by ‘mix’). B: The substrate preferences of FhcatB1 for the P2, P3 and P4 positions were determined using a combinatorial peptide inhibitor library and are displayed as the percentage of competition for each amino acid substituted at the indicated position. ‘n’ represents norleucine (a substitute for methionine). C: Data from positional scanning of the P2 position of FhcatB1 compared with other papain family proteases. Substrate preferences of the S2 subsite of papain family cysteine proteases were determined using combinatorial libraries. The preferences are presented in red for substrates with greater competition, or blue for those with little or no competition, with 50% competition in white. Amino acids, represented as single letters, are above the chart. The enzymes shown other than FhcatB1 are: CalpII, calpain II purified from porcine kidney (EMD Bioscience); CatH, cathepsin H purified from human liver (EMD Bioscience); CalpI, calpain I purified from porcine kidney (EMD Bioscience); CatC, cathepsin C purified from bovine spleen (Sigma); CatB, cathepsin B purified from bovine spleen (Sigma); CatF, recombinant cathepsin F; Papain, purified from Carica papaya (Sigma); CatS, recombinant human cathepsin S; Cruzain, cruzipain from Trypanosoma cruzi; CatL, recombinant human cathepsin L; rFalcipain 2, recombinant falcipain-2 from Plasmodium falciparum; CatK, recombinant human cathepsin K.