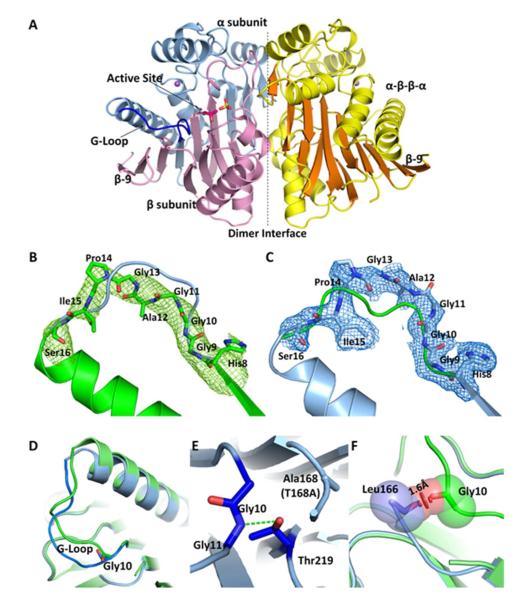
Figure 2.
A) The overall structure of hASRGL1 displays a conserved Ntn αββα fold with α-helices colored yellow and β-sheets colored orange. The dashed line indicates the interface between the dimers. The wild-type enzyme autocatalytically cleaves between Thr168 and Gly167 into α (light blue) and β (light pink) subunits. A sulfate group (yellow sticks) binds in each substrate binding pocket in proximity to the active site nucleophilic residue Thr168 (Thr168Ala) (hot pink sticks). A conserved Gly-rich loop (blue) is close to the active site and plays an important role in protein activation and catalysis. Plant-type asparaginases coordinate a sodium cation (purple sphere) in each monomer. B) The Fo-Fc omit map of cp-hASRGL1 Gly-rich loop (green mesh). The final refined cp-hASRGL1 (green) is well fitted in the cp-hASRGL1 Gly-rich loop omit map representing a dramatic orientation difference with the final refined hASRGL1-Thr168Ala Gly-rich loop model (blue cartoon). C) The Fo-Fc omit map of hASRGL1-Thr168Ala Gly-rich loop (blue mesh). It also shows changed orientation from final refined cp-hASRGL1 model (green cartoon). These two omit maps doubly confirm that the Gly-rich loops of cp-hASRGL1 and hASRGL1-Thr168Ala have different orientations, and that this difference is not due to model bias. □ level of omit map is set as 1.8 Å. D) Superposition of Gly-rich loops of hASRGL1-Thr168Ala (light blue) and of cp-hASRGL1 (green). The carbonyl group of Gly10 points in the opposite direction as shown by sticks. E) The hASRGL1-Thr168Ala amide group of Gly11 forms a hydrogen bond (green dashed line) with the proposed general base residue Thr219. F) The superimposed structure of hASRGL1-Thr168Ala (light blue) and cp-hASRGL1 (green), indicates that the residues preceding Thr168 (Leu166 and Gly167) of hASRGL1 would clash with the Gly-rich loop if the protein were to assume the open conformation observed in cp-hASRGL1. The red disk represents a significant Van der Waals overlay.