Figure 2.
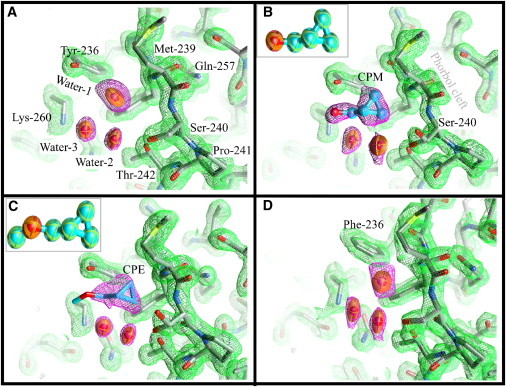
Details of the structure of the anesthetic binding site and the phorbol-binding cleft of the PKCδ C1B subdomain. Carbon atoms are colored gray, oxygen atoms red, nitrogen atoms blue, and sulfur atoms yellow. Thermal ellipsoids at 50% probability represent the water molecules. Waters in the phorbol cleft are omitted for clarity (see Fig. S2 for details). Ligand carbons are cyan. The 2Fo electron density map of the protein (green mesh) is drawn at 1σ cut-off. The equivalent maps around the ligands and water (magenta mesh) are at 1σ, except for CPE where the map is drawn at 0.9σ cut-off. (A) Wild-type C1B containing a water molecule (Water-1) at the anesthetic site. Water-2 and Water-3 form a hydrogen-bond network connecting Thr-242 and Lys-260. (B) CPM bound to C1B. CPM forms a hydrogen bond with Tyr-236. Water-2 and Water-3 form one side of the ligand-binding pocket. The hydroxyl of Ser-240 now adopts two rotamers. (Inset) Thermal ellipsoid representation of CPM. (C) CPM bound to C1B. The hydroxyl of Ser-240 again adopts two rotamers. (Inset) Thermal ellipsoid representation of CPE. (D) Structure of the C1B mutant Tyr-236-Phe. A water molecule occupies the anesthetic binding site in a way very similar to that observed for the wild-type. Neither CPM nor CPE were detected in this mutant (see text). Figures were prepared using Chimera (59).
