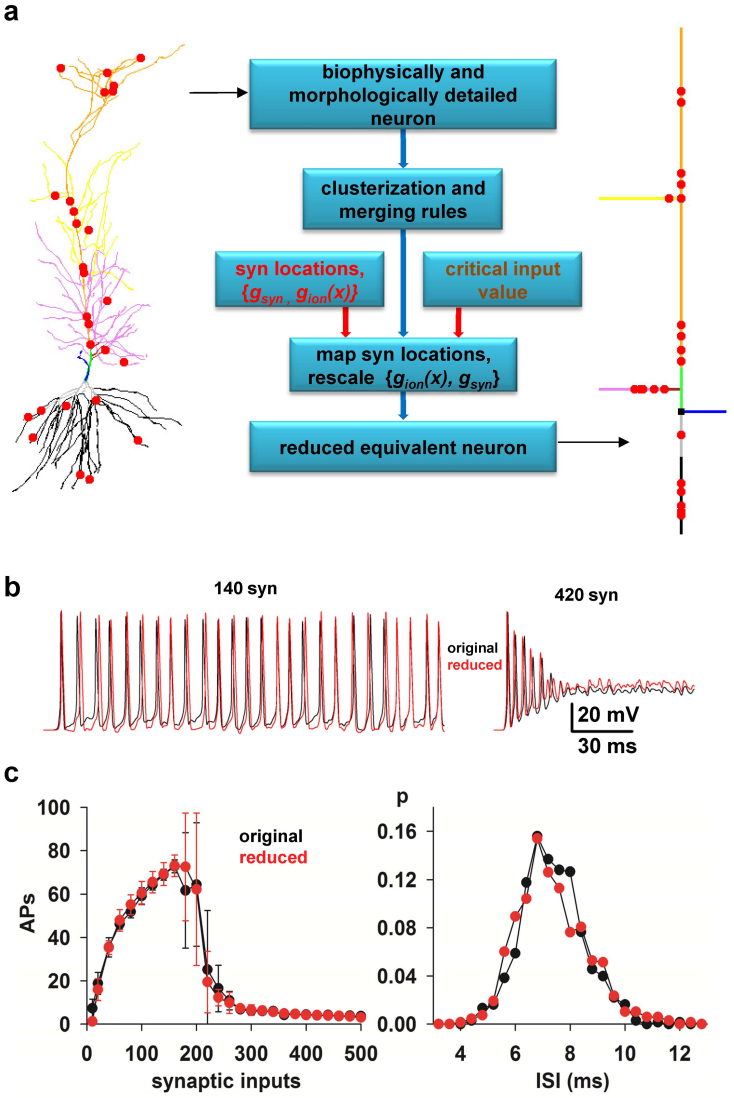
Figure 1. A method to reduce the computational complexity of realistic models of neurons.
(a) (left) typical 3D reconstruction of a realistic hippocampal CA1 pyramidal neuron (cell c70863 from the neuromorpho.org public archive), composed by 843 membrane segments; red circles indicate synapses location; different colors for dendrites highlight the different clusters used for this kind of cell population; (middle) Flow-chart illustrating the main steps to reduce a morphologically and biophysically detailed neuron model into a reduced, but functionally equivalent, version; (right) schematic representation of the equivalent model (27 membrane segments) obtained after application of the reduction method. (b) Somatic membrane potential of the full (black traces) and the reduced (red traces) model during a simulation activating 140 (left) or 420 (right) synapses. (c) (left) Average (n = 10, ±sd) number of APs elicited in 500 ms long simulations as a function of the number of synaptic inputs activated in the original (black) or in the reduced (red) model; the two curves are statistically indistinguishable (Wilcoxon Signed Rank test, p = 0.879); (right) normalized InterSpike Interval (ISI) distribution, from 10 simulations of the original (black) or the reduced (red) model during a simulation activating 140 synapses; the two curves are statistically indistinguishable (Wilcoxon Signed Rank Sum test, p = 0.626).