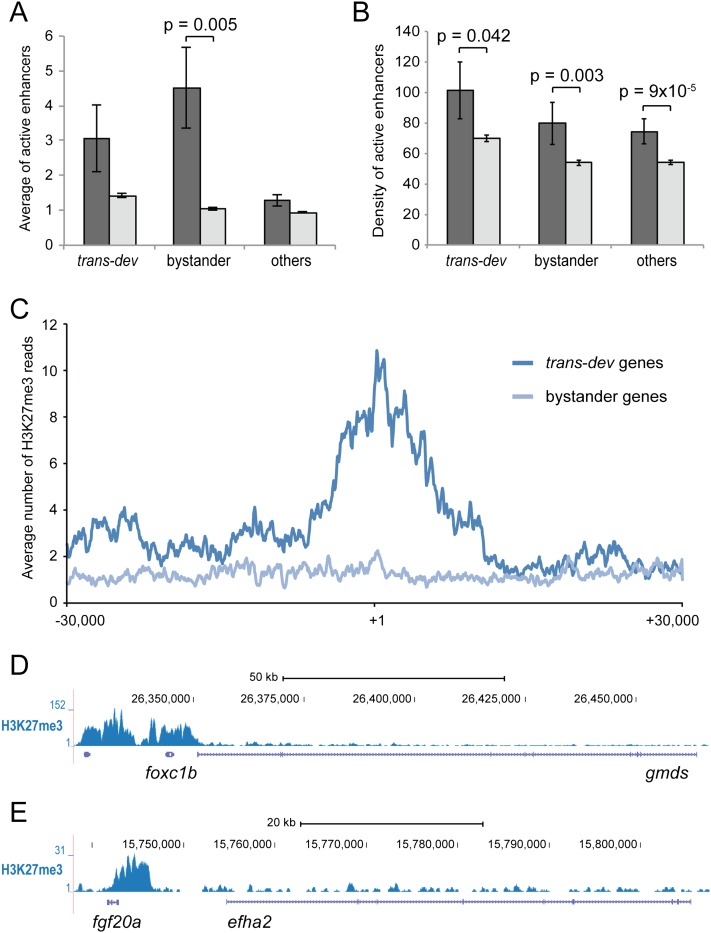
Figure 4.
Differential epigenetic marks in in bystander and trans-dev genes in early zebrafish development. (A,B) Average number (A) and density (peaks/Kbp, B) of H3K4me1+/H3K4me3− peaks (putative active enhancers) in 24-hpf zebrafish embryos within trans-dev, bystander, and other nondevelopmental genes for conserved pairs (dark gray) and for nonconserved pairs (light gray). Error bars correspond to standard errors. (C) Average number of reads for H3K27me3 ChIP-seq around transcription start sites show an specific deposition of this epigenetic mark in trans-dev genes. (D,E) Two examples of differential distribution of H3K27me3 in a trans-dev gene (foxc1b, D and fgf20a, E) and its nondevelopmental partner (gmds, D and efha2, E).