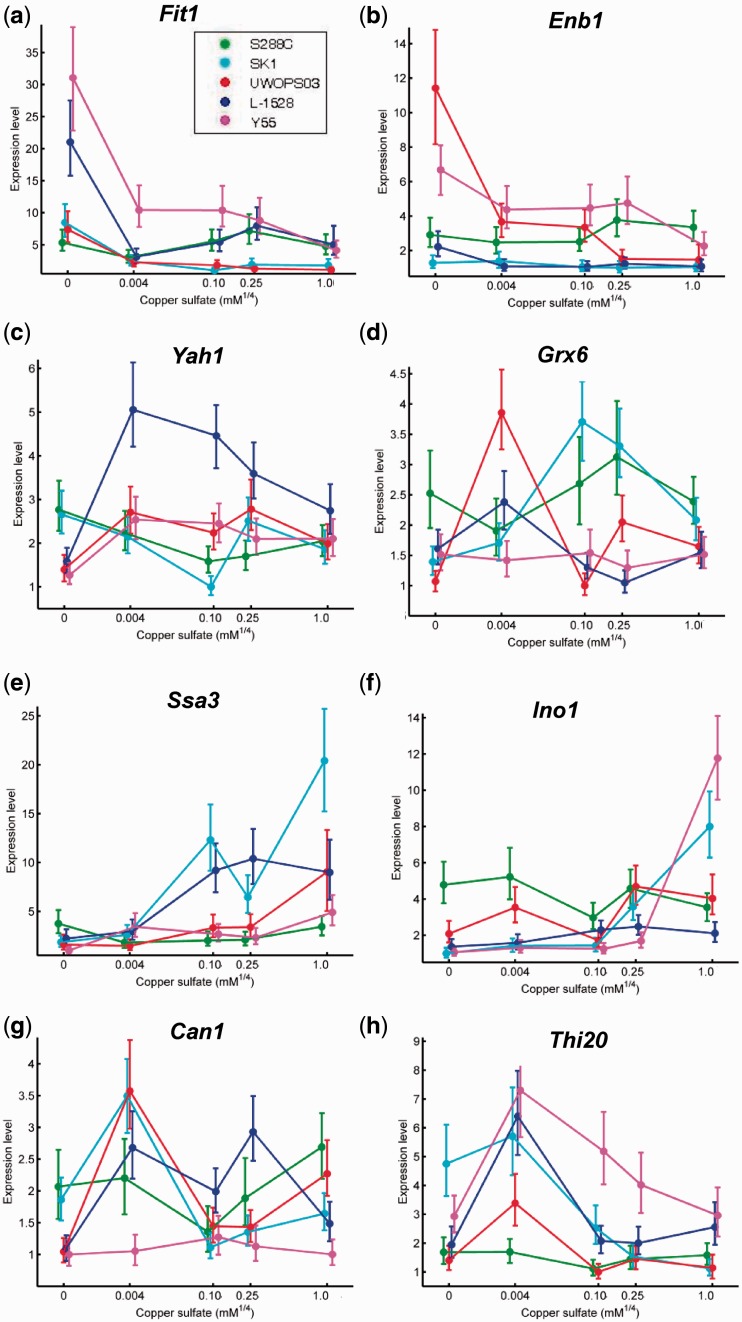
Fig. 7.—
Expression reaction norms for selected genes indirectly responding to copper. For each reaction norm, copper concentration is on the x axis, which is scaled by ¼ power to facilitate visualization of all five points on a convenient scale. The y axis is expression level relative to the lowest observed expression. Points plotted are the median of the posterior distribution. Error bars represent the 95% credible interval. Genes involved in copper-independent iron uptake (a) Fit1, encoding a siderophore-iron retaining mannoprotein, and (b) Enb1, encoding a ferric enterobactin transporter, exhibited strong upregulation of expression at low copper concentrations in some strains. (c) Non-essential gene Yah1, which encodes a mitochondrial ferredoxin and requires iron as a cofactor, exhibited marked downregulation in low copper in some strains. (d) Grx6, encoding a monothiol glutaredoxin that helps to cope with oxidative stress, exhibits diverse reaction norms in each strain, whereas oxidative stress gene (e) Ssa3, encoding an Hsp70-family ATPase, and membrane stress gene (f) Ino1, encoding a phospholipid bioynthesis gene, exhibit strong upregulation in high copper in some strains. Nitrogen catabolite repressed genes involved in amino acid biosynthesis are upregulated to differing degrees in differing strains in intermediate copper, including (g) Can1, encoding a plasma membrane arginine permease and (h) Thi20, encoding a protein functioning in thiamine metabolism.