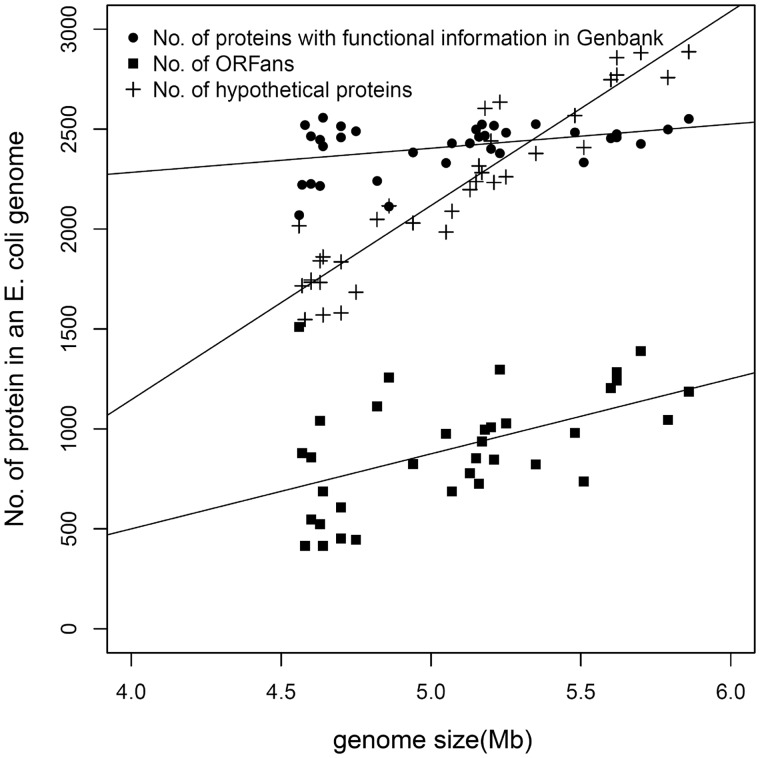
Fig. 3.—
ORFan composition as a function of genome size in 35 Escherichia coli strains. For each genome, counts are shown for three categories of putative protein-coding genes, along with regression lines. Two of the categories are mutually exclusive: each gene in a genome is either from a cluster (in the NCBI Protein Clusters database) that has a curated functional annotation (solid circle), or it is from a cluster annotated as “hypothetical protein” (plus symbols). The solid squares show the counts of ORFans, the vast majority of which are noncurated (see text). As genome size increases, the number of proteins with assigned functions remains nearly constant. The increase in genome size is not mainly attributable to ORFans, but is attributable to other genes for which functions are unknown.