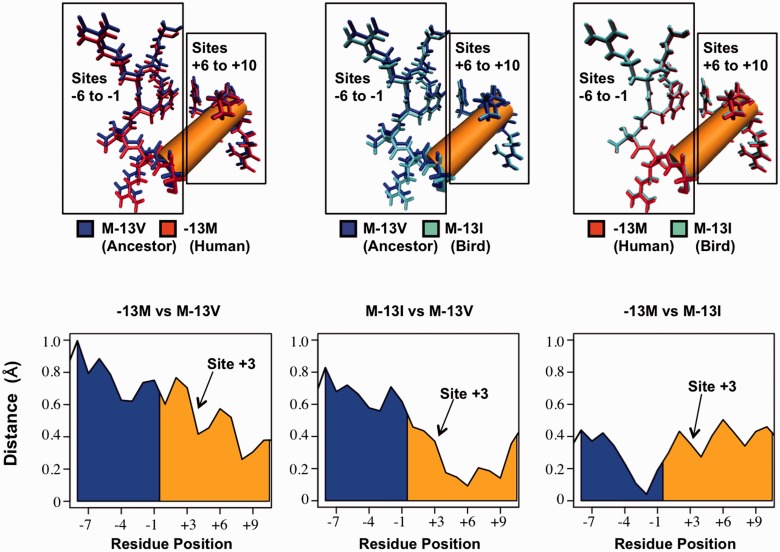
Fig. 4.—
Structural changes of SP1 zinc finger 2 (zf2) following replacements at site −13. (Top) Comparisons of predicted lowest-energy zf2 structures between the native human peptide (−13M), and peptides following replacements to the ancestral valine (M-13V) and bird isoleucine (M-13I) at site −13. Structural alignments were conducted according to residues on the 5′-end of the peptide (residues −16 to −12). Both −13M and M-13I peptides showed displacement of residues 5′ to the DNA-contacting alpha-helix (sites −6 to −1) compared with the ancestral valine peptide. No such displacement was seen between −13M and M-13I. All three peptides aligned closely at the 3′-end of the alpha-helix (sites +6 to +10), reflecting structural modifications at the 5′-end of the alpha-helix. (Bottom) Distances between alpha carbons prior to and within the alpha-helix (blue and orange, respectively). Comparisons between the native human peptide and M-13V (left) and between M-13I and M-13V (center) show closely aligned residues at the 3′-end of the alpha-helix and increasing displacement toward the 5′-end. These modifications begin around site +3, which directly contacts the A/C evolving site of the SP1-binding motif (Philipsen and Suske 1999; Bouwman and Philipsen 2002; Dhanasekaran et al. 2006). No such region-specific displacement between −13M and M-13I was observed between −13M and M-13I (right).