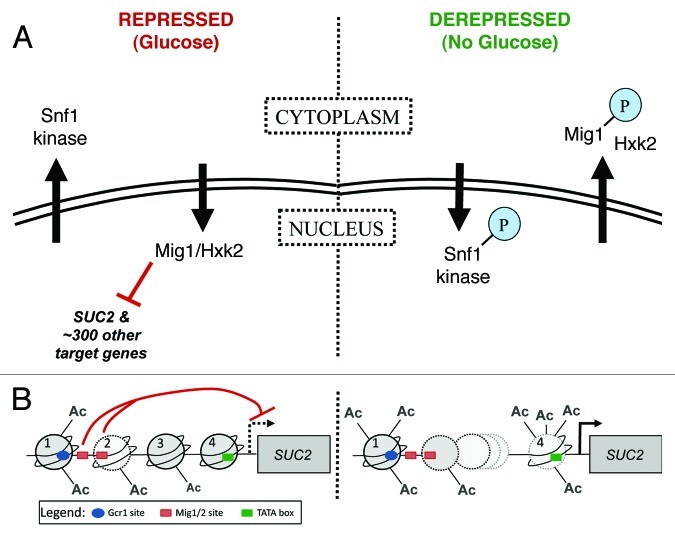
Figure 1. Regulation of glucose-repressed gene expression in Saccharomyces cerevisiae. Regulation of SUC2 expression is well characterized, and the proteins that carry out this regulation have been studied extensively. The SUC2 system therefore provides a good model for understanding how other genes controlled by the same factors are regulated. (A) A kinase-transcription factor pair work together to control expression of SUC2. Left panel. In the presence of glucose (repressed), the C2H2 zinc finger protein Mig1 binds DNA and inhibits transcription of SUC2, plus approximately 300 other genes. Complete inhibition also requires the hexokinase Hxk2. Snf1 kinase, a structural and functional homolog of mammalian AMPK, is found in the cytoplasm. Right panel. When glucose is depleted or withdrawn (derepressed), Snf1 is phosphorylated by upstream kinases, enters the nucleus and phosphorylates Mig1, which is then exported together with Hxk2. For detailed review, see reference 48. (B) Chromatin structure of the SUC2 promoter. Left panel. In the presence of glucose (repressed), four nucleosomes (numbered 1 through 4) cover the SUC2 promoter; a gradient of histone H3 acetylation decreases from 5′ to 3′.55 Two GC-rich sequences are required for repression of transcription (red t-bars) from the TSS (dashed black arrow).88 These sites are interchangeably bound in vivo by Mig1 and its homolog Mig2; function of the former, but not the latter, is regulated by the Snf1 kinase.48 The average location of the first Mig1/2 site (left-most red rectangle) is between nucleosomes 1 and 2. The second Mig1/2 site (right-most red rectangle) is close to the end of the DNA covered by nucleosome 289-91; nucleosome and repressor may compete for occupancy of this site. The TATA box (green rectangle) is covered by nucleosome 4.89-93 Right panel. Full induction of transcription requires the Swi/Snf chromatin remodeling complex.57,90,91,94-98 Swi/Snf associates more stably with promoter nucleosomes that have been acetylated by SAGA and NuA4 complexes57; in the absence of glucose, acetylation of H3 and H4 tails increases for all nucleosomes. The DNA formerly covered by two nucleosomes (nucleosomes 2 and 3) is more frequently covered by only one. Nucleosome 4, covering the TATA box, is hyperacetlyated and becomes unstable.55 Initiation occurs at the TSS (black arrow). Additionally required for full induction of SUC2 transcription are the nucleosome remodeling protein Spt699,100 and the transcriptional activator Gcr1 (blue oval, Gcr1 binding site).90,101-104 Nuclear pore proteins also interact with the SUC2 promoter13; it is important to learn how they collaborate with the well-characterized transcription factors described here.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.