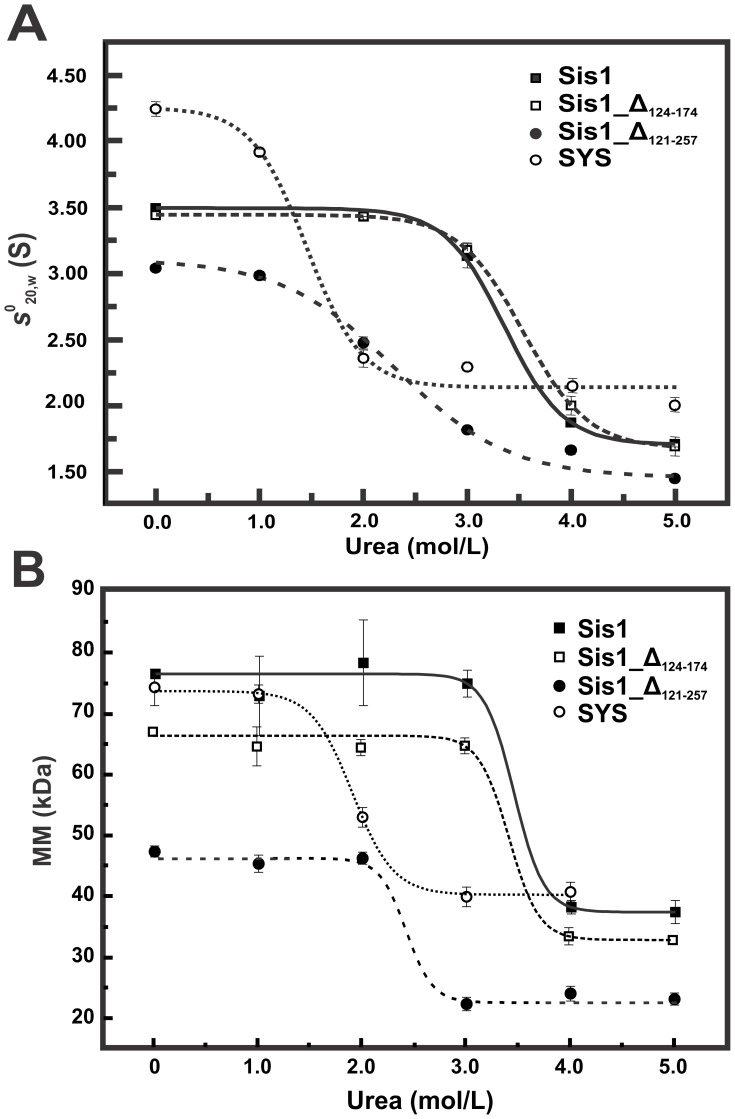
Figure 4. Urea disturbs the dimer structure of Sis1_Δ121–257 and SYS at lower concentrations than for Sis1 and Sis1_Δ122–174.
(A) SV-AUC experiments were done for all proteins (250–750 µg/mL) in the presence of urea at concentrations ranging from 1–5 M in buffer A. The curves of s 0 20,w versus urea concentrations were fitted by sigmoidal functions (continuous line) leading us to obtain the CmAUC (Table 1). Data at 0 M urea were obtained from [22] and [21]. (B) SEC-MALLS experiments were performed with all proteins in concentrations from 50 µM to 75 µM and in the presence of urea at concentrations ranging from 1–5 M in buffer A. The curves of MM versus urea concentration were fitted by sigmoidal functions (lines) resulting in the CmSEC-MALLS, which represents the urea concentration of the midpoint of the MM transition (Table 1). The molecular masses for wild-type Sis1, SYS, Sis1_Δ124–174 and Sis1_Δ121–257 were approximately 78, 75, 68 and 48 kDa in the absence of denaturant, and approximately 39, 42, 35 and 24 kDa in 4 M urea, respectively. Taken together, the results suggest that Sis1 and Sis1_Δ124–174 are dimers at higher urea concentrations than SYS and Sis1_Δ121–257.