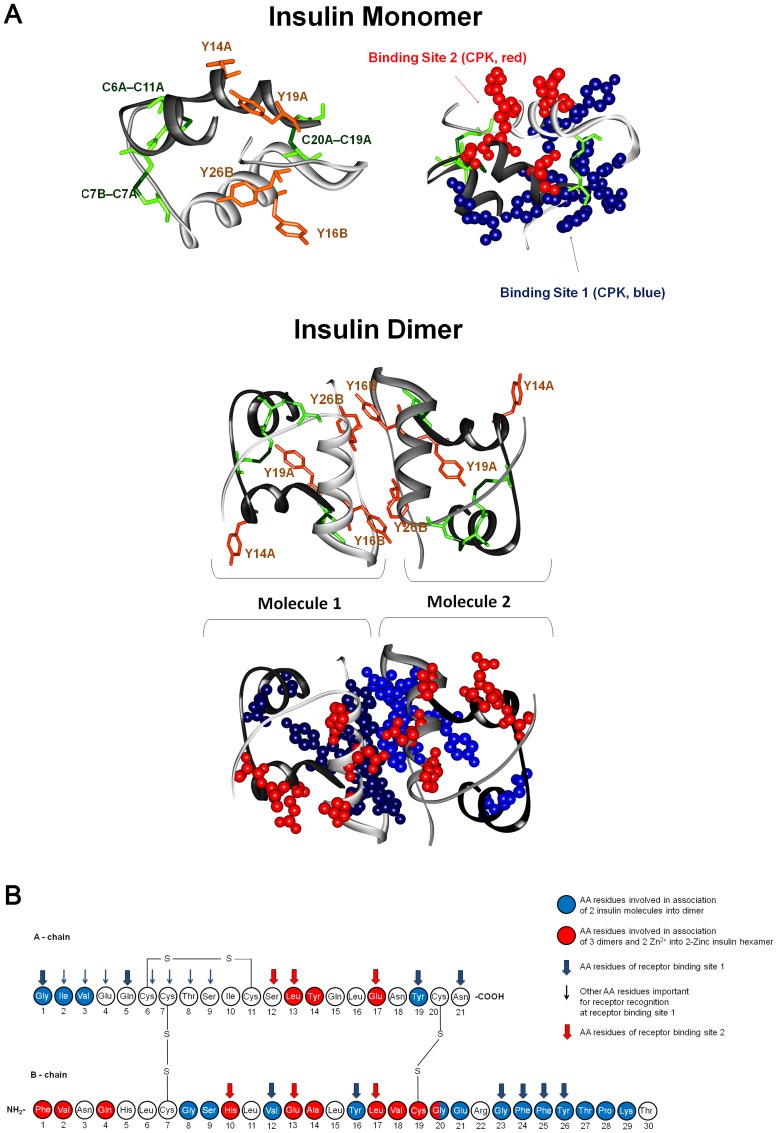
Figure 2. Tertiary and primary structures of insulin, dimer and 2Zn hexamer forms and receptor binding sites.
(A) 3D structures of insulin monomer and dimer extracted from the crystallized structured of the 2Zn pig insulin hexamer (4INS.pdb) [37]. Insulin A chains of insulin are displayed in gray (two different shades in the insulin dimer) and B chains are displayed in black. The 4 Tyr residues of insulin (Y, orange) and the 6 Cys (C, green) involved in 3 disulphide bridges (SS) are displayed in orange and green, respectively (left-side structure for insulin monomer and top structure for insulin dimer). The two insulin binding sites are displayed in CPK (0.5 Å radius) in blue (binding site 1) and red (binding site 2) (right side structure for insulin monomer and bottom structure for insulin dimer). (B) Primary structure of human insulin. The amino acid residues involved in the association of 2 insulin molecules into a dimer (blue residues) and in the molecular assembly of 3 dimers into 2Zn insulin hexamer (red residues) are displayed. The amino acid residues belonging to insulin receptor binding sites 1 and 2 are indicated by wide blue and red arrows, respectively. Amino acid residues important for insulin receptor recognition of the binding site 1 but not belonging to this site are displayed with thin blue arrows (adapted from [36], information regarding dimer association, hexamer formation, and insulin receptor binding sites extracted from [35], [36], [38], [48]–[51]).