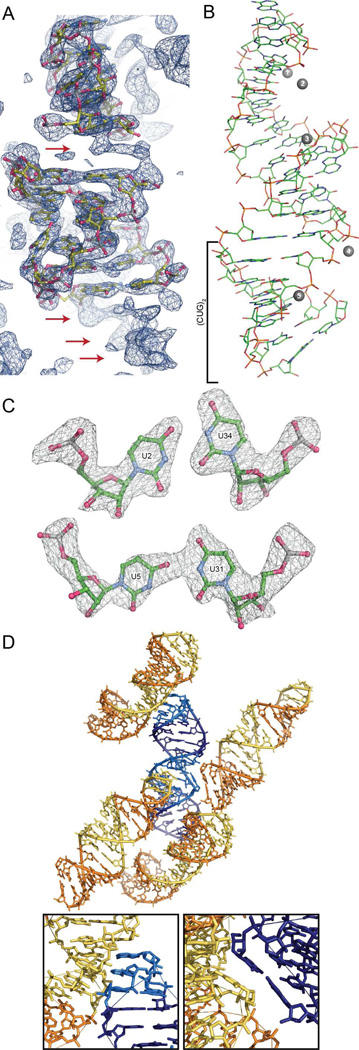
Figure 2.
Structure and crystal packing of trCUG-3. (A) Molecular replacement solution and 2Fo-Fc map (contoured at 1.5 σ) after one round of refinement. The density of the base pairs not yet modeled, both between the tetraloop/receptor and directly below the receptor, are clearly observable in the density (red arrows). (B) Tertiary structure of trCUG-3. The (CUG)2 region, containing four CUG repeats at the base of the hairpin, is denoted by a bracket. Magnesium ions are represented by numbered gray spheres. Waters are not shown. (C) Simulated annealing omit map (Fo-Fc) of both non-canonical U-U pairs contoured at 5 σ. (D) Crystal packing (or quaternary structure) of trCUG-3. Each RNA interacts with four other RNAs in the structure: at the loop, at the receptor, on the opposite side of the receptor and at the bottom of the helix. Tetraloop and receptor are denoted by lighter colors. Inset gives a closer view of each interaction.