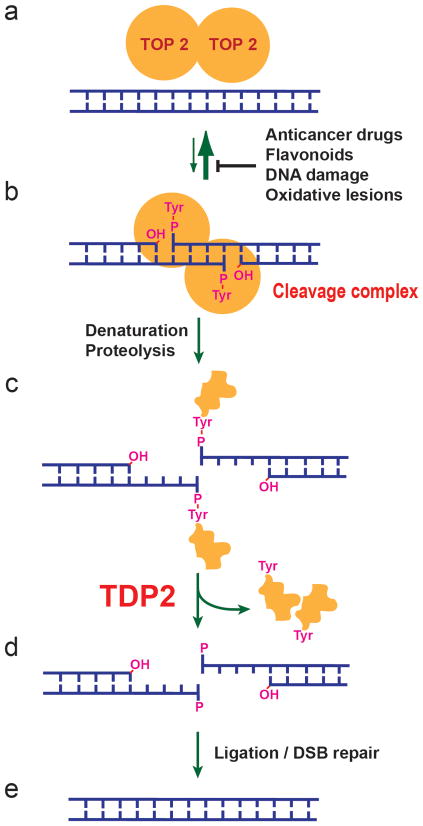
Figure 1. Repair of topoisomerase II cleavage complexes by TDP2.
(a, b) Topoisomerase II (TOP2) functions as a homodimer, where each monomer cleaves one strand of a double-stranded DNA by forming a covalent 5′-phosphotyrosyl bond. The resulting cleavage complex (b) allows the passage of another duplex DNA through the break, thereby enabling DNA relaxation, catenation-decatenation, and knotting-unknotting1. Under normal conditions, religation of the cleaved DNA strands is highly efficient and most of TOP2 is non-covalently bound to DNA as in (a). In the presence of anticancer drugs such as etoposide, mitoxantrone, doxorubicin, and daunorubicin, or food flavonoids or DNA damage or oxidative lesions, the cleavage complex accumulates and needs to be removed for DNA repair1. (c) Prior to TDP2 activity, the cleavage complex needs to be denatured or proteolyzed to expose the DNA-5′-phosphotyrosyl bond. (d) TDP2 releases the TOP2 polypeptide and leaves the 5′-phosphate end. (e) The DNA break may be repaired by direct ligation after annealing of the two ends with the 4-base pair stagger, or through double-strand break (DSB) repair mechanisms.