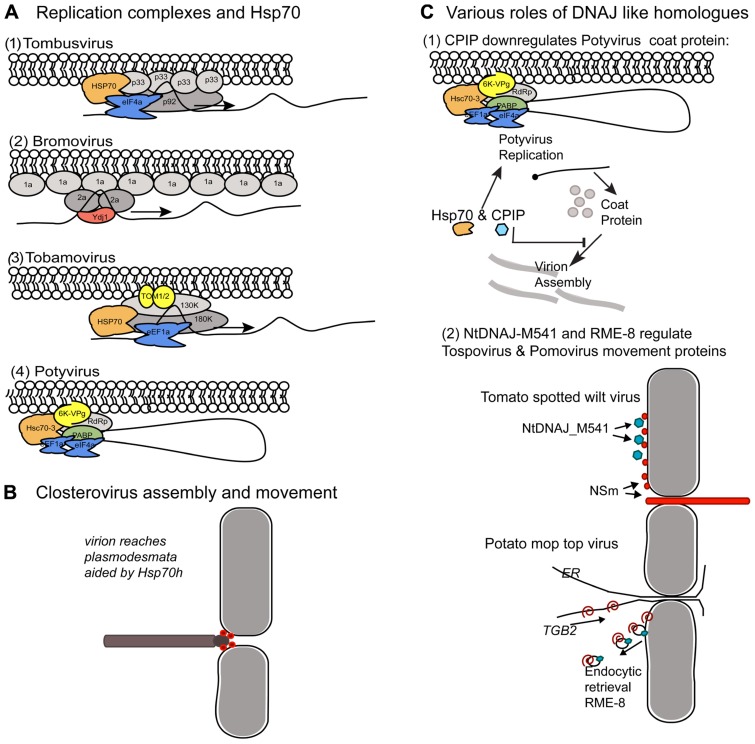
FIGURE 2.
Comparison of the host protein chaperones recruited to membrane bound viral complexes by unrelated (+) strand RNA viruses. Similar chaperones provide different roles in the viral protein complexes. (A) Comparison of Hsp70 and Ydj1 interactions with various viral replicases. (A1,2) The tombusvirus and bromovirus replicases assemble in spherules along the peroxisome and ER membranes, respectively. (A3,4) Tobamoviruses and Potyviruses replicate in ER derived structures. (A1) Tombusviruses encode p33 and p92 proteins required for replication. The Hsp70 (orange) recruits p33 to cellular membranes. eIF4e (blue) is another cellular component of the replication complex. (A2) Bromoviruses encode 1a and 2a proteins. 1a forms a shell along the membrane. Ydj1p is a J-domain protein involved in negative-strand synthesis which interacts with the 2a protein. (A3) Tobamovirus replicase consists of the 130K and 180K proteins and accumulates on ER membranes. TOM1 and TOM2 are host proteins which provide membrane anchoring. Hsp70 and eEF1a are host factors that associate with the viral replicase. (A4) The potyvirus replicase is anchored to ER membranes by the viral encoded 6K-VPg. The host PABP, eEF1a, eIF4a, and Hsc70-3 proteins associate with the viral replicase. It is likely that the PABP brings the 3′ end of the genome near the 5′ end and that replication is initiated along a circular RNA. (B) Closterovirus virions are long filamentous particles with structurally differentiated tail domain. The viral movement protein is an Hsp70 homolog (Hsp70h; red spheres) which functions to both stabilize the tail region of the virion and aid trafficking across plasmodesmata. (C) Role of DNAJ homologs in regulating virus encapsidation and egress. (C1) Depiction of the Hsc70-3 containing viral replicase and its relationship to another Hsp70 and CPIP protein. This describes another role for Hsp70 in the potyvirus life cycle, unlike its role in replication depicted in (A4). The Hsp70 and CPIP depicted here suppresses coat protein accumulation and blocks virion assembly. Virion assembly also serves to suppress viral genome translation and therefore, suppression of CP accumulation can enhance genome expression. This machinery reduces the impact of CP on viral genome translation. In this model, proposed by Hafren et al. (2010), CPIP recruits the potyvirus coat protein to Hsp70 which serves to aid ubiquitination and CP degradation. CPIP recruits the CP and thereby reduces its impact on viral genome translation. (C2) Yeast two-hybrid assays carried out using the tospovirus NSm protein identified NtDNAJ_M541 protein as an interacting partner. NSm (red spheres) associates with the plasma membrane, binds nucleocapsids, weakly binds RNA, and forms tubules. Given the myriad of NSm activities it is not yet clear how NtDNAJ_M541 (blue octagons) contributes to its functions. Pomovirus TGB2 movement protein is a transmembrane protein that resides in the ER and interacts with RME-8. TGB2 binds viral RNA and potentially cargoes it along the ER to plasmodesmata to facilitate intercellular transport. Researchers proposed that RME-8 is an endocytic marker indicating TGB2 is recycled from the plasma membrane back to the ER where it can bind viral genomes for further rounds of transport to the plasmodesmata.