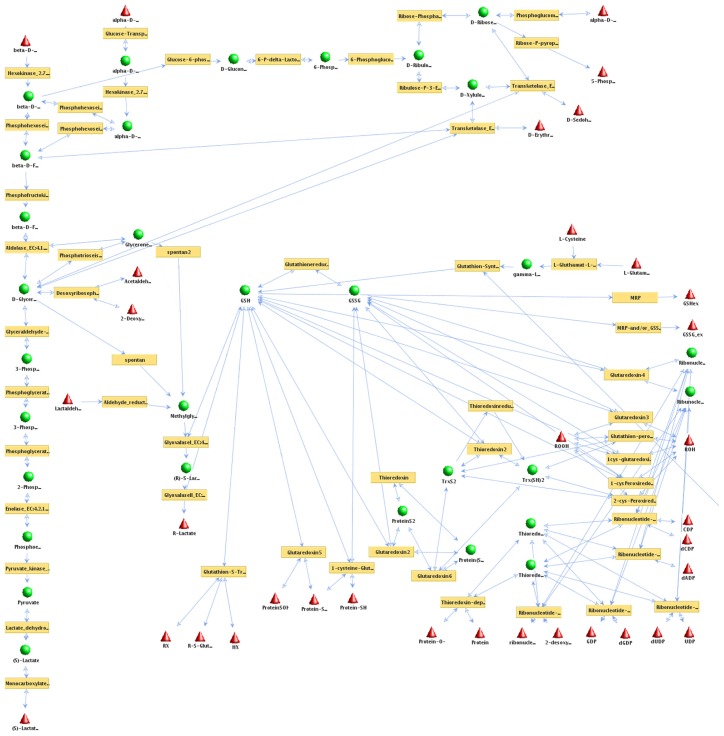
Figure 2.
Pathways of plasmodial redox metabolism.
Notes: All accessible pathways of the redox network from Plasmodium spp were calculated applying elementary mode analysis (software: YANA). Enzyme names are written in brown boxes. A metabolite (names in black) is either a substrate or a product of a metabolic reaction. External metabolites (red triangles): metabolite is either taken up from the environment (substrate) or constitutes the end product of an enzymatic reaction. These are the sources or drains of the metabolic fluxes modeled and hence need not be balanced. Internal metabolite (green balls): metabolite concentration has to satisfy the steady state condition of the model as it is produced to cover the need of reactions using it in turn again as a substrate. Each of these internal metabolites has to be balanced by the enzymatic reactions of the metabolic network. All pathways are shown as equally active (blue arrows indicate fluxes) as this calculation indicates only which pathways could be used at all. To avoid cluttering of the picture, some connections to external metabolites are not shown. For full detail on all enzymes, substrates and fluxes calculated see Additional files. No transcription data or experimental mRNA expression data were incorporated at this stage.