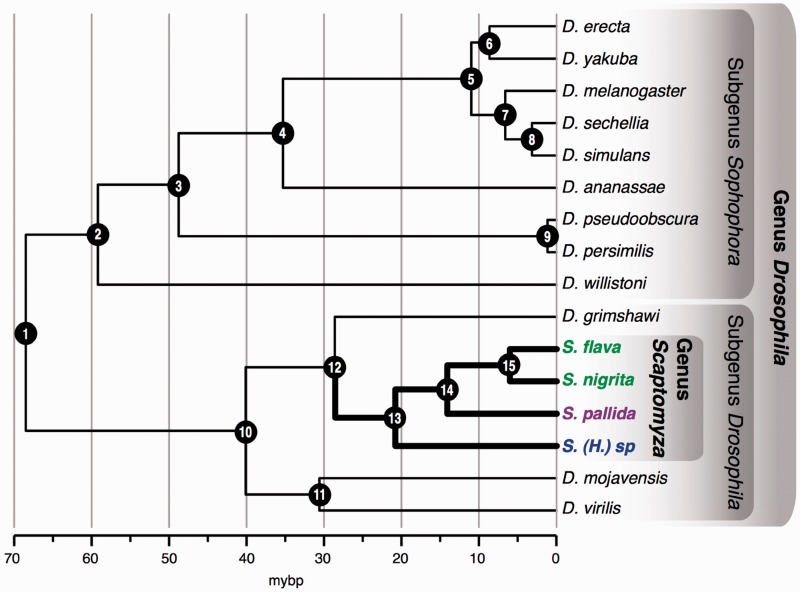
Fig. 1.—
Chronogram of combined nucleotide sequence data set from 9 genes from the 12 Drosophila species with completely sequenced genomes and multiple Scaptomyza species (S. flava, S. nigrita, S. pallida, and S. [Hemiscaptomyza] sp.). Initial tree was a maximum likelihood partitioned analysis of 9 genes inferred using RAxML (−ln L = −29,522.877). See table 1 for bootstrap support values for nodes. Nodes were constrained to ages estimated from Tamura et al. (2004). The two leaf-mining Scaptomyza species are resolved as sister taxa. Scaptomyza (Parascaptomyza) pallida and Scaptomyza (Hemiscaptomyza) sp. do not mine leaves of plants and can be reared on Drosophila media, and larvae are associated with decaying vegetation. The split between the most recent common ancestor of the two leaf-mining taxa, S. flava and S. nigrita (labeled in green), was estimated at 6.31(±0.63 SD) MYBP, and we inferred this to be the most recent date for the evolution of mustard leaf mining.